| droSim2 |
3r:13890810-13891155 - |
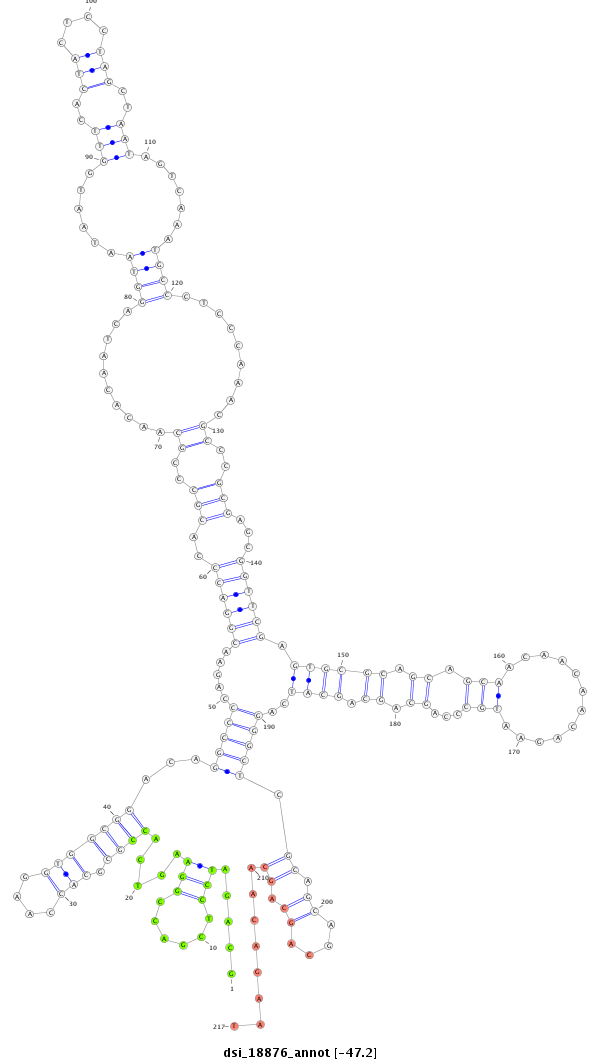
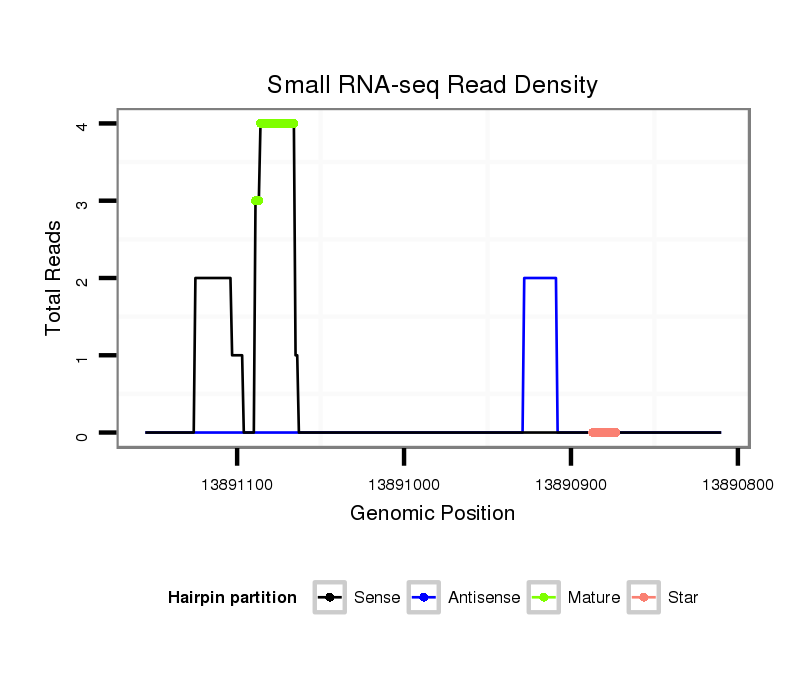
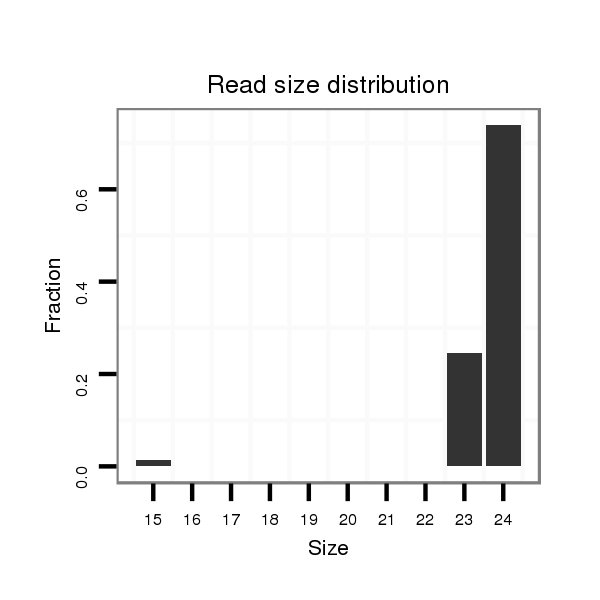
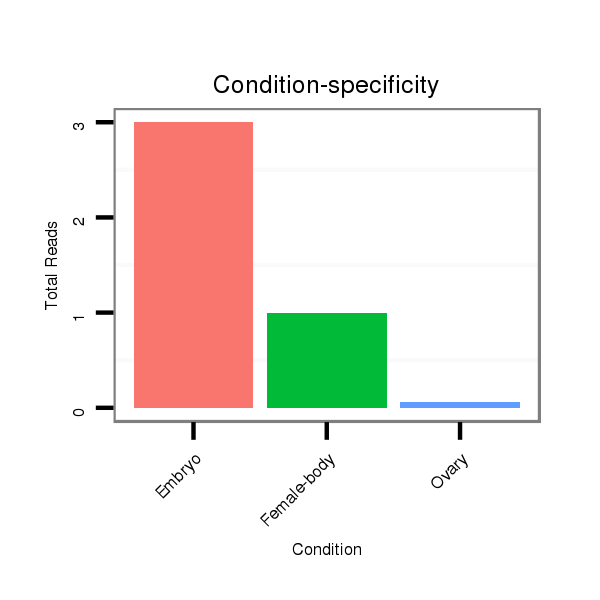
dsi_18876 |
CAGCAAGCAG--GCGGACTCCGTGACCGCGGCTGCCGAGCAGCTGAACAAGGACCTGCTGCTGGCTTCGCAGATCCTCGACCGGAAGTCACCGCGCACCAAGGTGG---CGGACAG----------------------------------------GCCC-CAGAACGGACCCA---CGCCCGCAACACAATCAGGTA---ATAATGGTTCACTACTCCTAGCTAATAGTCAAATGCCCTCCCAAACGC--CCG---CGAGCGG---TTC------------------------------GAGTGCGCAG-------------------------------------------------------CAGCA------------------------ACAACAACAGAA------------------------------------TGCCCAGCAGCAGCA---TCAGGGC---TCGCAG------------------------CA---------------------------------------------GCA------------------GCAGCA----------------------ACAGAATGTGGTGCAGCAGCAAAATGTGGCGCA---GCAGCAGCA------C---------------ATGC------AG------CAGCAAC------AGCAGC----------------------------AGTCGCAC---------- |
| droSec2 |
scaffold_0:14752903-14753248 - |
|
CAGCAAGCAG--GCGGACTCCGTGACCGCGGCTGCCGAGCAGCTGAACAAGGACCTGCTGCTGGCTTCGCAGATCCTCGACCGGAAGTCACCGCGCACCAAGGTGG---CGGACAG----------------------------------------GCCC-CAGAACGGACCCA---CGCCCGCAACACAATCAGGTA---ATAATGGTTCACTACTCCTAGCTAATAGTCAAATGCCCTCCCAAACGC--CCG---CGAGCGG---TTC------------------------------GAGTGCGCAG-------------------------------------------------------CAGCA------------------------ACAACAACAGAA------------------------------------TGCCCAGCAGCAGCA---TCAGGGC---TCGCAG------------------------CA---------------------------------------------GCA------------------GCAGCA----------------------ACAGAATGTGGTGCAGCAGCAAAATGTGGCGCA---GCAGCAGCA------C---------------ATGC------AG------CAGCAAC------AGCAGC----------------------------AGTCGCAT---------- |
| dm3 |
chr3R:7201415-7201766 + |
|
CAGCAAGCAA--GCGGACTCCGTGACCGCGGCTGCCGAGCAGCTGAACAAGGACCTGCTGCTGGCTTCGCAGATCCTCGACCGGAAGTCACCGCGCACCAAGGTGG---CGGACAG----------------------------------------GCCC-CAGAACGGACCCA---CGCCCGCAACACAATCAGGTA---ATAATGGTTCACTACTCCTAGCTAATAGTCAAATGCCCTCCCAAACGA--CCG---CGAGCGG---GTC------------------------------GAGTGCGCAG-------------------------------------------------------CAGCA------------------------ACAGCAACAGAA------------------------------------TGCCCAGCAGCAGCA---TCAGGGC---TCGCAG------------------------CA---------------------------------------------GCA------------------GCAGCA----------------------ACAGAATGTGGTGCAGCAGCAGAATGTGGCGCA---GCAGCAGCA------T---------------ATGC------AG------CAGCAGCAGCAACAGCAGC----------------------------AGTCGCAT---------- |
| droEre2 |
scaffold_4770:14411871-14412222 - |
der_628 |
CAGCAAGCAG--GCGGACTCCGTGACCGCGGCTGCCGAGCAGCTGAACAAGGACCTGCTGCTGGCTTCCCAGATCCTCGACCGGAAATCACCGCGCACCAAGGTGG---CGGACAG----------------------------------------GCCC-CAGAACGGACCCA---CGCCCGCAACACAATCAGGTA---ATAATGGTTCACTACTCCTAGCTAATAGTCAAATGCCCTCCCAAACGC--CCG---CGAGCGG---TTC------------------------------GAGTGCGCAG-------------------------------------------------------CAACA------------------------ACAGCAACAGAA------------------------------------TGCCCAGCAGCAGCA---TCAGGCC---TCGCAG------------------------CA---------------------------------------------GCA------------------GCAGCA----------------------ACAGAATGTGGTGCAGCAGCAGAATGTAGCACA---GCAGCAGCA------C---------------ATGC------AG------CAGCAGCAACAACAGCAGC----------------------------AATCGCAT---------- |
| droYak3 |
3R:11243292-11243646 + |
dya_1008 |
CAGCAAGCAG--GCGGACTCCGTGACCGCGGCTGCCGAGCAGCTGAACAAGGACCTGCTGCTGGCTTCGCAGATCCTCGACCGGAAATCGCCGCGCACCAAGGTGG---CGGACAG----------------------------------------GCCC-CAGAACGGACCCA---CGCCCGCAACACAATCAGGTA---ATAATGGTTCACTACTCCTAGCTAATAGTCAAATGCCCTCCCAAACGC--CCG---CGAGCGG---GTC------------------------------GAGTGCGCAG-------------------------------------------------------CAGCA------------------------ACAGCAACAGAATGC------AC---AGCAGCAGC---------------------AGCAGCA---TCAGGGC---TCGCAG------------------------CA---------------------------------------------GCA---------------------GCA----------------------ACAGAATGTGGTGCAGCAGCAGAATGTGGCGCA---GCAGCAGCA------C---------------ATGC------AG------CAGCAGCAACAACAGCAGC----------------------------AGTCGCAT---------- |
| droEug1 |
scf7180000409797:834646-834994 + |
|
CAGCAAACAG--GCGGACTCTGTGACTGCAGCGGCCGAACAACTAAACAAGGACCTGCTGCTGGCTTCGCAGATCCTCGACCGGAAGTCACCGCGCACCAAAGTAG---CGGATAG----------------------------------------GCCC-CAGAACGGACCCA---CGCCCGCAACACAATCAGGTA---ATAATGGTTCACTACTCCTAGCTAATAGTCAAATGCCCTCCCAAACGC--CCG---CGAATGG---GTC------------------------------GAGTGCGCAG-------------------------------------------------------CAGCA------------------------ACAGCAGCAGAA------------------------------------TGCTCAGCAGCAACA---TCAGAGC---TCGCAA------------------------CA---------------------------------------------GCA------------------GCAACA----------------------ACAGCAGCAAGCGCAGCAACAGAATGTGGCGCA---ACAACAGCA------T---------------ATGC------AG------CAGCAACAGC---AGCAGC----------------------------AATCGCAT---------- |
| droBia1 |
scf7180000302075:3552461-3552821 - |
|
CAGCAAGCAG--GCGGACTCCGTGACCGCGGCTGCCGAGCAGCTGAACAAGGACCTGCTGCTCGCCTCGCAGATCCTCGACCGGAAATCGCCGCGCACCAAGGTGG---CGGACAG----------------------------------------GCCC-CAGAACGGACCCA---CGCCCGCAACACAATCAGGTA---ATAATGGTTCACTACTCCTAGCTAATAGTCAAATGCCCTCCCAAGCGC--CTGTGTCGAGTGGTGGGTC------------------------------GAATGGGCAG-------------------------------------------------------CAGCAACAGCAGCAGCAGCA---------GCAGCAACAGAA------------------------------------TGCCCAGCAGCAACA---TGCGAGT---GCGCAG------------------------CA---------------------------------------------ACA------GCAACAG---CAGCAGC---AGACGT-------------------------CGC---A------------GCA---GCAACAGCA------C---------------CTGC------AA------CAGCAGCAGCAACAACAGC----------------------------AATCGCAT---------- |
| droTak1 |
scf7180000415763:230913-231243 - |
|
CAGCAAGCAG--GCGGACTCCGTGACGGCGGCAGCCGAGCAACTGAACAAGGACCTGCTGCTCGCCTCGCAGATCCTCGACCGGAAATCGCCGCGCACCAAGGTGG---CGGACAG----------------------------------------GCCC-CAAAATGGACCCA---CGCCCGCAACACAATCAGGTA---ATAATGGTTCACTACTCCTAGCTAATAGTCAAATGCCCTCCCAAACGC--CCGTCGCGAGTGG---CTC------------------------------GAGTGGGCAGCAGCAGCAG-------------------------------------CA------------------AC------------------------AGAC---------GTCGCAA---------------------------CAGCAACA-TGTTAG--TAACGTG---------------------------------------------------------------------------CA---ACAACAACAG---ACGCCGCA----------------------A------------------------------CA---GCAGCAACA------T---------------TTGC------AG------CAGCAACAGCAA---CCAC----------------------------AGTCGCAT---------- |
| droEle1 |
scf7180000491080:2493022-2493391 - |
|
TAGCAAGCAG--GCGGACTCCGTGACGGCGGCCGCCGAACAGCTGAACAAGGACCTGCTGCTGGCCTCCCAGATCCTCGACCGCAAGTCGCCGCGCACCAAGGTGG---CGGACAG----------------------------------------GCCC-CAGAACGGACCCATCGCGCCCGCATCACAATCAGGTA---ATAATGGTTCACTACTCCTAGCTAATAGTCAAATGCCCTCCCAAACGC--CCG---CCAGCGG---GTC------------------------------GAGTGGGCAGCAACAGCAG-------------------------------------CA------------------GCAGCAGCAGCAACAGCAACAGCAACAGAA------------------------------------TGCGCAGCAGCAACA---TCAGGTC---GCGCAG------------------------CA---------------------------------------------GCA------ACAACAG---CAGCCGCA----------------------ACA---------GCAGCAGCAGAGTGTGGGGCAACCGCAGCAGCA------C---------------CTGC------AG------C------------AGCAGC----------------------------AATCGCAT---------- |
| droRho1 |
scf7180000779097:111018-111366 - |
|
CAGCAAGCAG--GCGGACTCCGTGACCGCGGCGGCCGAGCAGCTGAACAAGGACCTGCTGCTCGCTTCGCAGATCCTCGACCGGAAATCGCCGCGCACCAAGGTGG---CGGACAG----------------------------------------GCCC-CAGAACGGACCCA---CGCCCGCATCACAATCAGGTA---ATAATGGTTCACTACTCCTAGCTAATAGTCAAATGCCCTCCCAAACGC--CCG---CGAGTGG---GTC------------------------------GAGTGGGCAG-------------------------------------------------------CAGCAACAGCAAC---------------------AGCAGAA------------------------------------TGCACAGCAGCAACA---TCAGGTA---TCGCAG------------------------CA---------------------------------------------GCA------GCAACAG---CAGC-------------------------------------CGCAGCAGCCGAATGTGGCGCA---ACAACAGCA------T---------------TTGC------AG------CAGCAGCAGCAACAGCAGC----------------------------AATCGCAT---------- |
| droFic1 |
scf7180000453912:1686710-1687067 - |
|
TAGCAAGCAG--GCGGACTCGGTGACCGCAGCGGCGGAGCAGCTGAACAAGGACCTCCTGCTGGCATCGCAGATCCTCGACCGGAAGTCGCCGCGCACCAAGGTGG---CGGACAG----------------------------------------GCCG-CAAAACGGACAAA---CGCCCGCAACACAATCAGGTA---ATAATGGTTCACTACTCCTAGCTAATAGTCAAATGCCCTCCCAAACGA--ACG---CGAGCGG---TTC------------------------------GAGTGCGCAG-------------------------------------------------------CAGCAACAGCAGC---------------------AGCAGAATGC------AC---AGCAGCAGCAGCAGCAACAC------------------------AGC---TCGCAG------------------------CA---------------------------------------------AC---------AACAG---TCGCAGCA----------------------ACAGCA------GCCACCGCAAAATGTGTCGCA---GCA---ACA------C---------------ATAC------AA------CAGCAACAGCAGCAACAGC----------------------------AATCGCAT---------- |
| droKik1 |
scf7180000302276:549546-549894 - |
|
CAGCAAGCAG--GCGGACTCCGTGACGGCAGCGGCCGAGCAGCTCAACAAGGACCTGCTGCTGGCCTCGCAGATCCTCGACCGGAAGTCGCCGCGCACCAAGGTGG---CGGACCG----------------------------------------GCCC-CAGAACGGACCCG---CGCCCGCAACACAATCAGGTA---ATAATGGTTCACTACTCCTAGCTAATAGTCAGATGCCCTCCCAACCGC--CGG---CGAGCGG---GGC------------------------------GAGTGCGCAGCAGCAGCA----------------------------------------------------------------------------------GCAGAATGC------GC---CGCAGCAGCAGCAGCCACCAAGTGCCCAACAACAACA---TCCGGGC---TCCCAG------------------------CA---------------------------------------------GCA------------------GC----------------------------------------AGCAGCAAAACGCAGTGCA---GCAACAGCA------C---------------TTGC------AG------CAGCA---------GCAGC----------------------------AAGCGCAT---------- |
| droAna3 |
scaffold_13340:10624102-10624426 - |
|
CAGCAAGCAG--GCGGACTCCGTGACCGCAGCCGCCGAACAGCTGAACAAGGACCTGCTGCTGGCATCACAGATCCTGGACAGGAAATCGCCGCGCACCAAGGTGG---CAGACCG----------------------------------------GCCC-CAGAACGGACCAA---CACCCGCATCCCAATCAGGTA---ATAATGGTTCACTACTCCTAGCTAATAGTCAGATGCCCGCCCAGCCGT--CCG---CG------------------------------------------AATGCGCAGCAGCAGTTG-------------------------------------CA------------------GC------------------------AGGC---TCAGGGATCCCAGCAGCAACAGCAGCAGCCAGGATCGCAA---------------------------------------------------CA---------------------------------------------GC---------CACAGCCAAG----------------------TTCGCAGCA---------GCAGCAGCAGAATGTGGTGCA------------------------------------------------------------------GCAAC----------------------------AGCCGCAT---------- |
| droBip1 |
scf7180000396374:452324-452648 + |
|
CAGCAAGCAG--GCCGACTCCGTGACCGCAGCGGCCGAGCAGCTGAACAAGGACCTGCTGCTAGCCTCCCAGATCCTGGACAGGAAATCGCCGCGCACCAAGGTGG---CAGACCG----------------------------------------GCCC-CAGAACGGACCCA---CACCCGCATCCCAATCAGGTA---ATAATGGTTCACTACTCCTAGCTAATAGTCAGATGCCCGCCCAGCCGT--CCG---CG------------------------------------------AATGCGCAGCAGCAGCTG-------------------------------------CA------------------GC------------------------AGGC---TCAGGGATCCCAGCAGCAACAGCAGCAGCCAGGATCGCAA---------------------------------------------------CA---------------------------------------------GC---------CACAGCCAAG----------------------CTCGCAGCA---------GCAGCAGCAGAATGTGGCGCA---GCA---------------------------------------------------------------AC----------------------------AGCCGCAT---------- |
| dp5 |
2:29212726-29213093 - |
dps_862 |
CAGCAAGCAG--GCGGACTCGGTGACAGCCGCCGCCGAGCAGCTCAACAAGGACCTGCTGCTGGCATCGCAGATACTCGACCGCAAGTCGCCGCGCACCAAAGTGG---CGGACAGAGGAGGA----------------------------CCCGGAGCC-CAGAACGGCCCCA---CGCCGACCACACAATCAGGTA---ATAATGGTTCACTACTCCTAGCCCCTAGTCAAATGCCCCCCCAACCGC--C-G---CCAGCGGTG-GTCGTG-------------------------GCCAATGCCCAG-------------------------------------------------------CAGCAGCAGCAACAGCAGCA---------GCAGCAGCAGAA------------------------------------CGTGCAGCA-------------------------------------------------------------------------------------------------------------------------------------------------------------GCAGCAGCAGAATGTGCCACA---GCAGCAGCAGCAGCAGGCACCGCAACAGAATCCCC------AG------CAGCAAC------AGCAGC----------------------------CGCCGAATGTGCAGCATC |
| droPer2 |
scaffold_1383:9455-9762 - |
|
CAGCAGGCGGTGGCACAGGCGGCGGCGGTGGCGGTGGACC-----------GCCCGGCTCTCCGCTCTGCAGCTCCCCGTCCAGC-GTCACAGCATC-----GCAG---CAGGCCACAG---TCGCCGCAGTAGCAGCCGCCACATTTAATGGAAGTGCCACA---------------GATAGCAGCAT-GTCGGGCA---ACA------CATCACCGATAGCCAGCGGCGAGCCGCTGCTCCAGACGC--CGC---CC------------------------------------------GCCATGCAACAGCAGCAG-------------------------------------CA------------------GC---A------------ACAAC---AG---------------CAGCAACAA---------------------------------------------CAG------------------------CA---------------------------------------------GCA---GCAACAACAG---CAGCAGCA----------------------A------------------------------------CAGCAG---------------------------CAGCAGCAG------------CAGCAACAACAGC----------------------------AGCAGCAG---------- |
| droWil2 |
scf2_1100000004902:4277696-4278062 - |
dwi_2476 |
TAGCAAGCAA--GCGGACTCGGTGACGGCAGCAGCAGAGCAGCTCAACAAGGATTTGTTATTGGCTTCCCAGATACTCGACCGGAAATCGCCCCGTACCAAAGTAG---CCGATCGCAGCGGA---------CACAGCGGCA-------ATACTGGCATC-CAGAATGGCCCCA---CGCCGGCAACACAATCAGGTA---ATAATGGTTCACTACTCCTAGCTAATAGTCAAATGCCCTCCCAACCGC--CTA---CGGGCAA---CAA------------TCTTGCCGGTAATCCGAACAATGTGCAACAACAGCAG----------------------------------------------CAACA------------------------ACAGCAACAAAA------------------------------------TGCACAGCAACAACA---CCCATCG---TCGGCA------------------------CA---------------------------------------------GCA------------------GCCGCA----------------------ACA------------TCAGCCAAGTGTTCCCCA---GC---------------------------------------------------CAG------CGCCGC----------------------------CTCCACAT---------- |
| droVir3 |
scaffold_13047:3247771-3248158 - |
dvi_15378 |
CAGCAAACAG--GCGGATTCGGTAACGGCGGCCGCCGAGCAGCTCAACAAGGATCTGATGCTGGCCTCCCAGATACTCGATCGCAAATCGCCGCGCACCAAAGTCGTCGCCGATCGCGG---ACTCAGCGGCAACAGCGGCC-------ATCCGGGCGCC-CAGAACGGTCCGA---CGCCAGCAACACAATCAGGTA---ATAATGGTTCAATACTATTAGCTAATAGTCAG------------------------CAGGCGA---GTCAGGGTC------------------------A---GGGCAACAGCAGTA------------------GTGCTGTCAATGCAAATCAGCAGCAGCAGCAGCA------------------------ACAACAACAGCA------------------------------------------------------TCAACTT---GCACAG---------------------CAGCA------------------------------------GCAGCA---GCA------------------GCAACAACAGACCGCACAGCAGCA-CCAACA---------------------------GCA---ACAGCAGCA------A---------------CAAC------AG------CAGCAAC------AGCAGC----------------------------AACAACAG---------- |
| droMoj3 |
scaffold_6540:20288401-20288779 - |
dmo_2001 |
CAGCAAGCAG--GCGGACTCGGTGACCGCAGCTGCCGAGCAGCTCAACAAGGATCTGATGCTGGCCTCCCAGATACTCGATCGCAAGTCGCCGCGCACCAAGGTCGTCGCGGATCGCGG---CCACGGCGGCAGCAGCAGCAACGCC-AACTCGGGCGCC-CAGAGCGGATCGG---CATCGGCCATACAATCAGGTAACAATAACGCCTCACTACTACTAGCTAATAGTCAGC------------------AG---CAAGCGA---GTCAGGGTC------------------------A---GGGCAACAGCAGCAG-------------------------------------CA------------------GC------------------------AGCA---------------------------------------CCAGCAGCAGCACTGTCAG--T---CAACAGCAACAGCAGCAACTTGCACAG---CA---------------------------------------------GCA-------------------------------------------------------GCAGCAGCAGCAACAGGCCGCACA---ACAACAGCA------C---------------CAAC------AG------CAGCAAC------AGCAAC----------------------------AGCAGCCT---------- |
| droGri2 |
scaffold_15074:7620694-7621157 - |
dgr_254 |
CAGCAAGCAG--GCGGATTCAGTGACCGCAGCAGCGGAGCAGCTCAACAAGGATCTGATGTTGGCCTCGCAGATACTCGACCGGAAGTCGCCGCGAACGAAGGTCGTCGCGGATCGGGG---A----------------------------ACGGGCGCC-CAAAATGGTCCGA---CGCCAGCAACACAATCAGGTA---ATAATGGTTCACTACTACTAGCTAATAGTCAAATGCCCGCACAGCAGCAGCAG---CAGGCAA---GTCAGAGTCAAAGTC------------------A---AAACAGCAACAGCAGCAGCAGCAACAACAGTAATGTTGTC------------------------------------------------------------AA------------------------------------TGCACAGCAGCAGCA---ACAGC---------------AGCAGCAACTTGCACAACAGCAGCAGCATAGCGCACAGCAGCAACTTGCACAGCAACAACAACAGCAGCAGCAACAACAACAG---CAGCAGCA----------------------A------------------------------------CAGCAG---------------------------CAGCAGCAGCAGCAACAGCAGCAGCAA---CAACAGCCGCAACTAAACCAATCACAAGTCATGTCTGCAA---------- |