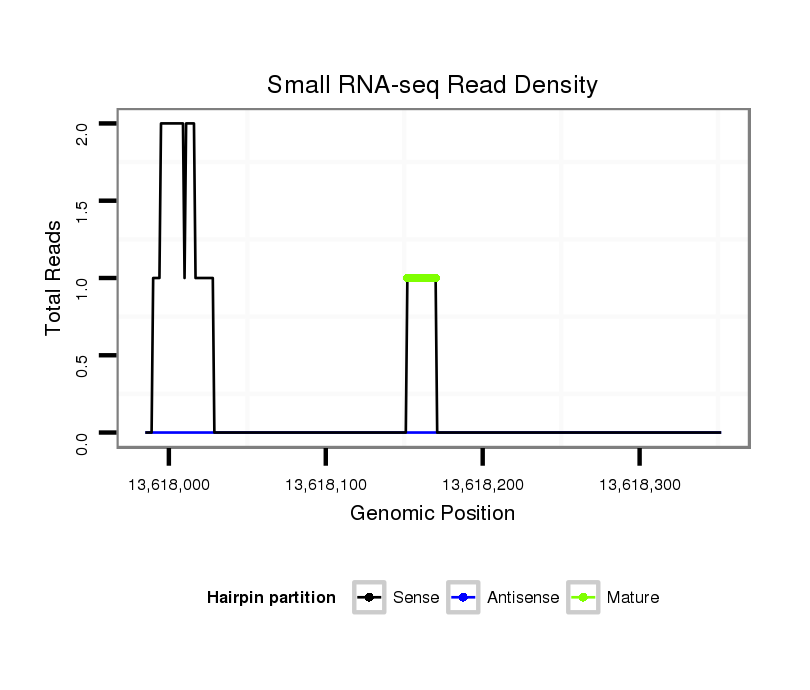
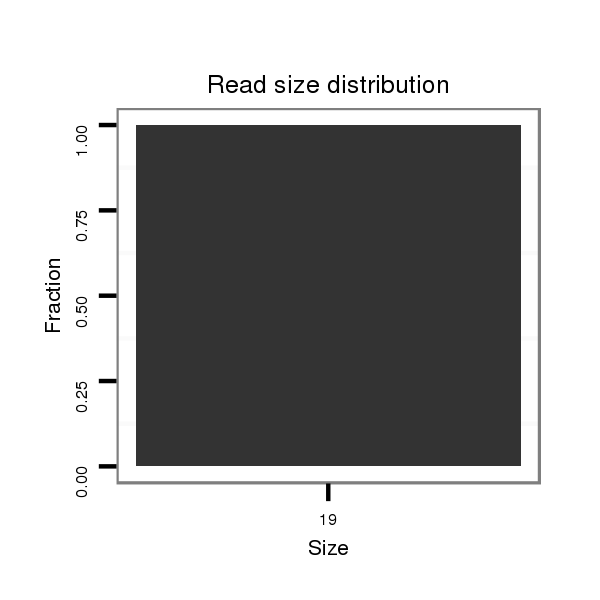
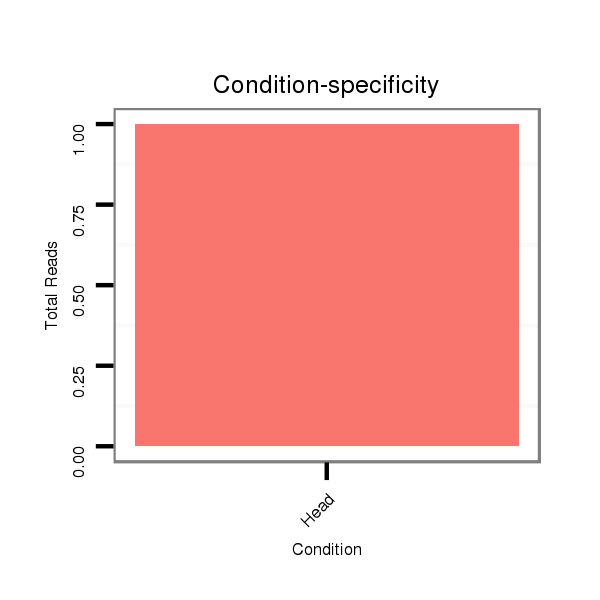
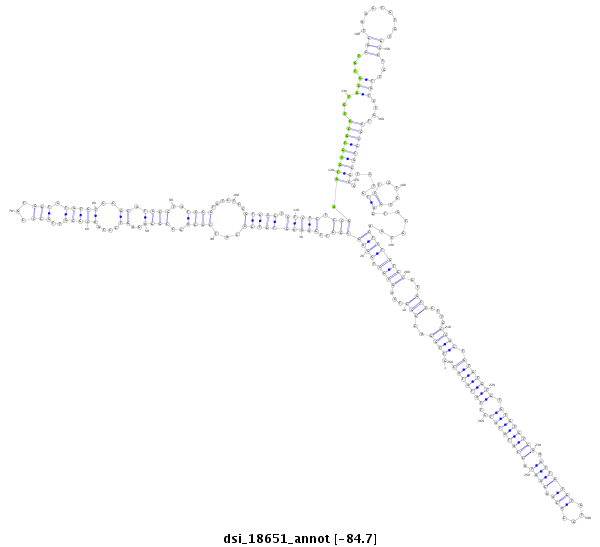
| droSim2 |
2l:13617985-13618352 + |
dsi_18651 |
TTGCCTTTT-------TGCACA------GCGGCTGAGC-----GATTGAGCGATTGAACGATTGA--------ACGGTTGGACGGCT------------------GAGCGCTGGAGCGGTTGAGAGTG-TG-CCATC-----GGAGCCGGAGAGT---------------CCTAACGCGTTGCCCGTGGTC------------------------TG------TGTCCGCATCCGTAT----------------------------------------C----------CGTATCTCC--GTG-----------------CGTCTCTCTCCGGGTGTGTG--CGT---------------------------------------------------GT--------GCGTGTG----G------C------------TA------------------AGTG----T------GTGCCTGTTGTCTGTC-G-------------CGCGCGC--G-------------------------TC--TGTCTGT--GTGAGTGTGTG-ACTCT------------GTGCGTGCCC---TTGCCAGTGTGTGTGTGT------GTGT-GGAATTGT-GTGTGCAC-------AACAATACCAT------------------ATATCT-------------------------------------------------TATATA-------CTAATACCAGTGT--GCCCACACTCGCACACGGCC----------TGCAAA---TAAAACATTAGA |
| droSec2 |
scaffold_3:298547-298923 + |
|
TTGCCTTTT-------TGCACA------GCGGCTGAGC-----GATTGAGCGATTGAACGATTGA--------ACGGTTGGACGGCT------------------GAGCGCTGGAGCGGTTGAGAGTG-TG-CCATC-----GGAGCCGGAGAGT---------------CCTAACGCGTTGCCCGTGGTC------------------------TG------TGTCCGCATCCGTAT----------------------------------------C----------CGTATCTCC--GTG-----------------CGTCTCTCTCCGGGTGTGTG--CGT---------------------------------------------------GT--------GCGTGTG----G------T------------TA------------------AGTG----T------GTGCCTGTTGTCTGTC-GC----TAACGGTGCGCGCGC--G-------------------------TC--TGTCTGT--GTGAGTGTGTG-ACTCT------------GTGCGTGCCC---CTGCCAGTGTGTGTGTGT------GTGT-GGAATTGT-GTGTGCAC-------AACAATACCAT------------------ATATCT-------------------------------------------------TATATA-------CTAATACCAGTGT--GCCCACACTCGCACACGGCC----------TGCAAA---TAAAATATTAGA |
| dm3 |
chr2L:14040227-14040609 + |
|
TTGCCTTTT-------TGCACA------GCGGCTGAGC-----GATTGAGCGATTGAACGATTGA--------ACGGTTGGACGGCT------------------GAGCGCTGGAGCGGTTGAGAGTG-TG-CCATC-----GGAGCCGGAGAGT---------------CCTAACGCGTTGCCCGTGGCC------------------------TG------TGTCCGCATCCGTAT----------------------------------------C----------CGTATCTCC--GTG-----------------CGTCTCTCTCCGGGTGTGTG--CGT---------------------------------------------------GT--------GCGTGTG----G------T------------TA------------------AGTG----T------GTGCCTGTTGTCTGTC-GC----TAACGGTGCGCGCGC--G-------------------------TC--TGTCTGT--GTGAGTGTGTGGACTCT------------GTGCGTGCCCCC-CTGCCAGTGTGTGTGTGTG----TGTGTGGGTTTTGT-GTGTGCAC-------AACAATACCAT------------------ATATCT-------------------------------------------------TATATA-------CTAATACCAGTGT--GCCCACACTCGCACACGGCC----------TGCAAA---TAAAATATTAGA |
| droEre2 |
scaffold_4929:11429538-11429918 - |
|
TTGCCTTTT-------TGCACA------GCGGCTGAGC-----GATTGAGCGATTGAACGATTGA--------ACGGTTGGACGGCT------------------GAGCGCTGGAGCGGTTGAGAGTG-TG-CCATC-----GGAGCCGGAGAGT---------------CCTAACGCGCTGCCCGTGGTC------------------------TG------TGTCCGCATCCGTAT----------------------------------------C----------CGTATCTCC--GTG-----------------CGTCTCTCTCCGGGTGTGTG--CGTGTGCG---------------------------------------------TGTG--------CGTGTG----G------T------------TA------------------AGTG----T------GTGCCTGTTGTCTGTC-GC----TAACGGTGCGCGCGC--G-------------------------TC--TGTCTGT--GTGAGTGTGTG-ACTCT------------GTGCGTGCCC---CTGCCAGTGTGTGTGTGCG----TGTGTG-----TGT-GTGTGCAC-------AACAATACTAT------------------ATATCT-------------------------------------------------TATATA-------CTTATACCAGTGT--ACCCACAATCGCACACGGCC----------TGCAAA---TAAAATATTAGA |
| droYak3 |
2L:14133316-14133707 + |
|
TTGCCTTTT-------TGCACA------GCGGCTGAGC-----GATTGAGCGATTGAACGATTGA--------ACGGTTGGACGGCT------------------GAGCGCTGGAGCGGTTGAGAGTG-TG-CCATC-----GGAGCCGGAGAGT---------------CCTAACGCGCTGCCCGTGGTC------------------------TG------TGTCCGCATCCGTAT----------------------------------------CCGTATCT---CGTATCTCC--GTG-----------------CGTCTCTCTCCGGGTGTGTG--CGTGTGGT---------------------------------------------TGTG--------CGTGTG----T------T------------TA------------------AGTG----T------GTGCCTGTTGTCTGTC-GC----TAACGGTGCGCGCGC--G-------------------------TC--TGTTTGT--GTGAGTGTGTG-ACTCT------------GTGCGTGCCC---CTGCCAGTGTGTGTGTGTGTGCGTG--TG-----TGT-GTGTGCAC-------AACAATACTAT------------------ATATCT-------------------------------------------------TATATA-------CTAATACCAGTGTGTGCCCACACTCGCACACGGCC----------TGCAAA---TAAAATATTAGA |
| droEug1 |
scf7180000409452:604266-604657 + |
|
TTGCCTTTTGCCTCCATGCACA------GCGGCTGAGC-----GATTGAGCGATTGAACGATTGA--------ACGGTTGGACGGTT------------------GAGCGGTGGAGCGGTTGAGAGTGGCA-CCATC-----GGAGCCGGAGAGC---------------CCTGTCGCGTTGCCCGTGGTC------------------------TG------TGTCCGCATCCGTAT----------------------------------------C----------CGTAACTCC--GTG-----------------CGTCCCTCTCCGGGTGTGTGTGCGTCTTCG---------------------------------------------TGT--------GCGTGTG----C------C------------TA------------------TGTGT--GT----T-ATGCCTGTTGTCTGTC-GC----TAACGGTGCGCGCGC--G-------------------------TC--TGTCTGT--GTGAGTGTGTG-ACTCT------------GTGCGTGCCC---CTGCCACTGTGTTTGTGT------A--TG-----TGT-GTGTGCAC-------AACAATACTAT------------------ATATCT-------------------------------------------------TATATA-------CTAATACAAGTGTG-GCCCACACTCACACACGGCC----------TGCAAA--ATAAAATAGTAAA |
| droBia1 |
scf7180000302422:7769541-7769937 + |
|
TTGCCTTTTGCCTCCATGCACA------GCGGCTGAGC-----GATTGAGCGATTGAACGATTGA--------ACGGTTGGACGGTT--------GGACGG--TTGAGCGGTGGAGCGGTTGAGAGTG-TG-CCATCGTTGCGGAGCCGGAGAGT---------------CCTGTCGCGTTGCCCGTGGTC------------------------TG------TGTCCGCATCCGTAT----------------------------------------C----------CGTTTCTCC--GTG-----------------CGTCTCTCTCCGGGTGTGTG--CGTGTGCC---------------------------------------------TGTGCGTGT--GCGTGTGCC--C------C------------TG------------T------GTG----T------GTGCCTGTTGTCTGTC-GC----TAACGGTGCGCGCGC--G-------------------------TC--TGTCTGT--GTGAGTGTGTG-ACTCT------------GTGCGTGCCC---CTGCCAGTGT------------------G-----TGT-GTGTGCAC-------AACAATACTAT------------------ATATCT------------------------------------------------CTATATA-------CTAATACCAGTGTG-GACCACACTCACACACGGCC----------TGCAAA---CAAAGTATTAGA |
| droTak1 |
scf7180000415399:1266335-1266714 - |
|
TTGCCTTTTGCCTCCATGCACA------GCGGCTGAGC-----GATTGAGCGATTGAACGATTGA--------ACGGTTGGACGGTTGAGCGGTTGAGCGG--TTGAGCGGTGGAGCGGTTGAGA------------------------GAGAGT---------------CCTGTCGCGTTACCCGTGGTC------------------------TG------TGTCCGCATCCGTGT----------------------------------------C----------CGTATCTCC--GTG-----------------CGTCTCTCTCCGGGTGTGTG--CGTGTCCCTGTG--------------C--------------------GTGA-GTGT--------GCGTGTG----C------C------------TA------------------TGTGT--GTGTGCCTG-GCCTGTTGTCTGTC-GC----TAACGGTGCGCGCGC--G-------------------------TC--TGTCTGT--GTGAGTGTGTG-ACTCT------------GTGCGTGCCT--------------------------------------CT-GTGTGCAC-------AACAATACTAT------------------ATATCT------------------------------------------------CTATATA-------CTAATACAAGTGTG-GCCCACACTCACACACGGCC----------TGCAAA---TAAAATATTAGA |
| droEle1 |
scf7180000491338:1670068-1670429 + |
|
TTGCCTTTTGCCTCCATGCACA------GCGGCTGAGC-----GATTGAACGATTGAACGGTT----------------GGACGGTT--------G----G--TTGAGCGGTGGAGCCGTTGAGAGTG-TG-CTAAC-----GGAGCTGGAGAGC---------------TTAGCCGCGCTGCCCGTGGTC------------------------TG------TGTCTGCGTCTGTGT----------------------------------------CCGTGTCCGTGCGTATCTCC--GTG-----------CGTGTGCGTCTCTCTCCGAGTGTGTGTGCGTGTGCC---------------------------------------------TGTG--------CG------------------------------------------T------GTG----T------GTGCCTGTTGTCTGTC-GC----TAACGGTGCGCGCGC------------------------------------TG--------------------------------------------------------T------------G--TG-----TGT-GTGTGCAC-------AACAATACTAT------------------ACATAT------------------------ATATATATATATCTATATATATATATATATA-------CCAATACAAGTGT--GACCGCACTCACACACGGCA----------AGCAGA---TAATTTATTT-- |
| droRho1 |
scf7180000762282:51506-51907 - |
|
TTGCCTTCTGCCTCCATGCACA------GCGGCTGAGC-----GATTGAGCGATTGAACGATTGAGCGGTTGAACGGTTGGACGGTT--------GGACGGTGTTGAGCGGTGGAGCCGTTGAGAGTG-TG-CCATC-----GAAGCCGGAGAGT---------------CCTGTCGCGCTGCCCGTGGTC---------TGAG---------CCTG------TGTCCGCATCCGTGT----------------------------------------CCGTGTCCGTGCGTATCTCC--GTG-----------CGTGTGCGTCTCTCTCCGGGTGTGTGTGCGTGTGCC---------------------------------------------TGTG--------CG------------------------------------------T------GTG----T------GTGCCTGTTGTCTGTC-GC----TAACGGTGCGCGCGC--G-------------------------TC--TGTCTGT--GTGAGTGTGTG-ACTCT------------G----------------------------------------------TGT-GTGTGCAC-------AACAATCATAC----ACACAC--------AT------------------------------------------ATATATATATATATATA-------CTAATACAAGTGTG-GACCGCACTCACACACGGCC----------AGCAAA---TAAAATATTTAA |
| droFic1 |
scf7180000454082:3718-4159 - |
|
TTGCCTTTTGCCTCCATGCACA------GCGGCTGAGC-----GATTGAGCGATTGAACGATTGA--------ACGGTTGGACGGTTGAGCGGTTGGACGG--TTGAGCCGTTGAGCGGTTGAGAGTT-TG-CCACC-----GGAGCCGGAGAGT---------------CCTGTCGCGTTACCCGTGGTC------------------------TG------TGTCCGCATCCGTAT----------------------------------------C----------CGTATCTCCGTGTG-----------------CGTCTCTCTTCGGGTGTGTGAGTGTGTGCCTGTGGCTGT------------------GCGTTT--GCTT----GTTGTG--------CGTGTG----C------G------TTTGCGTTT------------------GCGTTGGT------GTGCCTGTTGTCTGTC-GC----TAACGGTGCGCGCGCTCG-------------------------TC--TGTCTGT--GTGAGTGTGTG-ACTCT------------GTGCGTGCCCCAGCTGCCACTGCGT------------G--TG-----TGT-GTGTGCAC-------AACAATACTAT------------------ATCCAT------------------------------------------------ATATATACTAAT-ACCAATACAAGTGTG-GCCCGCACTCACACACGACC----------TGCAAA---TAAAATATTTGA |
| droKik1 |
scf7180000301334:62292-62703 - |
|
CTGCCTTTTGCCTCCAAGCAC---------GGCCGAGA-----GATTGAGCGACTGAAC---------------------------T------------------GAGCGATTGAACGATTGAGAGTG-TG-CCAGC-----AGAGCCGGAGAGTCGCGTCGCGTTGCGTTGCGTTGCGTTGCCCGTGGTC------------------------TG------CGTCCGCATCCGCAT-CCTCCGAATCCTTGCCCGCCTTCGCCTCCGCCTCCGCCTCCGTGTCCGTG------------TG-----------------------------------------------------------------CTCCTCTC-CTCTCCTCGCCCATCC-G------TTT--CGCCCCG------------------------TTTGTG--------T------GCG----T------GTGTCTGTCGTCTGTC-GC----TAACGGTGCGCGCGC--G-------------------------TA--TAGCTGT--GTGAGTGTGCG-AGTTA------------GTGCGTGGCC---ATG--------T------------C--TG-----TGT-GTGTGCAC-------AACAATTCTAT------------------ATATAT----------------------------------------GTATACGTGTATATA-------CTAATACTAGCGT--GCCCACACTCACACAAG-CA-A--AAAGACTGCAAA---TAAAAAAT---- |
| droAna3 |
scaffold_12916:11739808-11740203 - |
|
TTGCCTTCTGCCTCCATGCACA------GCAGCTGAGG-----GATTGAGCGGTTGAACGGTT--------------------------------------------------GAGCGGTTGAGAGTG-TGTCCATC-----AGAGTCGG-GAGT---------------CGCGGCGCGTTGCCCGTGGTCGCATTCGCATAAGCATTCGCATCCCG------TATCCGTATCCGTCT----------------------------------------CCGTATCCGTG------------TGTCCT--------------------------GTGAGTG--CGTGTACTT------------------------------------------GCCC----C--CAGTGTGTG----T--------------------GTGTG--------T------TTG----T------GTGCCTGTTGTCTGTCTGT----TGGCGGTGCGCGCGC--G-------------------------TC--TATCTGTGTGTGAGTGTGTG-ACTCT------G-----GTGCGTGCCC----TG-------------------------G-----TGT-GTGTGCAC-------AACAACACTATCTATACATACATATTTATATACAT------------------------------------------------ATCTATGTGTATA-ATAAAAGAA------------AAACACACACAGGCAACAAAAAACAGCAAATAATAAAATAACAGA |
| droBip1 |
scf7180000396572:1405821-1406220 - |
|
TTGCCTTCTGCCTCCATGCACA------GCAGCTGAGG-----GATTGAGCGGTTGAACGGTT--------------------------------------------------GAGCGGTTGAGAGTG-TGCCCATC-----AGAGTCGG-GAGT---------------CGCGGCGCGTTGCCCGTGGTCGCATTCGCATAAGCATTCGCATCCCG------TATCCGTATCCGTCT----------------------------------------CCGCATCCGTG------------TGTCCT--------------------------GTGAGTG--CGTGTACATGTGCCTGTG----------------T---------------------------GCCCCTGCG----AGTG--TG------------TG------------T------GTG----T------GTGCCTGTTGTCTGTCTGC----TGGCGGTGCGCGCGC--G-------------------------TCTGTGTCTGT--GTGAGTGTGTG-ACTCT------------GTGCGTGGCC----TG---G----T------------G--TG-----TGT-GTGTGCAC-------AACAACACTATCTATACATACATATCTA------C-------------------------------------------------TATATATGTATAACCAAAAAAAG------------CACACACACAGACTAAAAAAAATATTAAAAAATAAAATAACAAA |
| dp5 |
4_group4:5625646-5625957 - |
|
TTGCCTTTT-TCTCCGTGCACA------GCCACCGAGA-----GATTGAG-------------------------------------------------------------------------------TT-CCATC-----AG---CGTAGTGT---------------CGCGTCGCGTTGGTTGTGGTC------------------------TG------TGTCCGTGTCAGTGT----------------------------------------C----------CGTATCTGT--GTGTGCTGCTCGAGCGTGTG----------CGTGAGTG----CG--------------TG----------------C---------------------------GAGCGTGTG----TGTG--TG------------TG----TGTGCCTCTGCCTGTGCC----T------GTGCCTGTTGTCTGTC-GC----TAACGGTGCGCGCGC--G-------------------------TG--TATCGGC--GAGAGTGTGTG-----C------------GT-----------------------------------GC--G-----TGTACTGTGCAC-------TAAAATGCCAT------------------GTACA--------------------------------------------------TAGATA-------CTAATACATGCCT---ACCACACACATACA-GACA----------TGGGAA---TAAA---TTAGA |
| droPer2 |
scaffold_16:1324142-1324447 + |
|
TTGCCTTTT-TCTCCGTGCACA------GCCACCGAGA-----GATTGAG-------------------------------------------------------------------------------TT-CCATC-----AG---CGTAGTGT---------------CGCGTCGCGTTGGTTGTGGTC------------------------TG------TGTCCGTGTCAGTGT----------------------------------------C----------CGTATCTGT--GTGTGCTGCTCGAGCGTGTG----------CGTGAGTG----CG--------------TG----------------C---------------------------GAGCGTGTG----TGTG--TG------------TG----TGTGCCTC------TGCC----T------GTGCCTGTTGTCTGTC-GC----TAACGGTGCGCGCGC--G-------------------------TG--TATCGGC--GAGAGTGTGTG-----C------------GT-----------------------------------GC--G-----TGTACTGTGCAC-------TAAAATGCCAT------------------GTACA--------------------------------------------------TAGATA-------CTAATACATGCCT---ACCACACACATACA-GACA----------TGGGAA---TAAA---TTAGA |
| droWil2 |
scf2_1100000004511:11008677-11008898 - |
dwi_741 |
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCCCGGGGTA------------------------AG------TGTTCTTTTCTGTGT----------------------------------------A----------TGTGT---G--GTG-----------------CGTCTGTGTGTGTGTGTGTG--TGTGTATG---------------------------------------------TGTG--TGT--GTGTGTATGTGTGTGTGTGTGTATGTGTGTGTGTGTG--------T------TTG----T------GTGC----------------------------G-------------------------------------------T--GTGTGTGTGTG-TTTGT------------GTGCGTGTGT---GTGTGTGTGTGTGTGTGT------GTGT-GGAGCTG---TATGTATAAACCAAAACAAATTTAT------------------ATATAT------------------------------------------------ATATATA-------TATATATTA--------------------------------------------------------- |
| droVir3 |
scaffold_12723:1119839-1120142 + |
|
TTGCCTTTTC-----CTGCACAGCCAA---AGCAGAGCTAAGCG-----------------------------------------------------------------------------------------------------------GAGA---------------CGCGTCGCG---GTCGTG-CC------------------------TG------TGTCTTTGTCTGTGT----------------------------------------T----------TGTGACTTGGACTG-----------------TGACTGTGTATGGAAGCGCGTGC-----------------------------------------------------GT--------GCGTGTG----T------G------------TA------------T------GTG----T------GTGTTTGTTGTCTGTC-GTCGCTTAGCACTGCGCGCGC--GTTTTGTCAGTGTATGTGTG------CT--TGTTTGT--ATGTGTGTGCG-CGTGTGCCCGTGATTCTCTGTGCTTCT---CTGTGCATATGTGTGTGT----------G-----TGC-GTGTGCGT-------GTTAAAAC----------------------------------AGTGTGACCATATAT----------------AT--------ATATATATA-----CATATA------------------------------------------------------------ |
| droMoj3 |
scaffold_6500:10254103-10254446 - |
|
TTGCCTTTTCCC----TGCACAGCCAAAGCAGCAGAGCCAAGCG-----------------------------------------------------------------------------------------------------------AAGT---------------CGCGTCGCG---GTCGTG-CC------------------------TG------TGTCTGCGTT----T----------------------------------------TTGTGAC----TGTGACTTT---------------------------------GCGTGCGTG--TGTGTGCTT----------------------------------GCTTATTT-G------TAT--GTGTGTG----T--G--CG------------TG------------T------GTG----TGTGTCAG-TTTTGTTGTCTGTC-GTCGCTTAACACTGCGCGCGC--GATTTGTCAGTGTATGTGTGCGAGCGTGCGTATGTGT--GT----------------------------------------GTGTGAGTGAGTCTGTGTG----CG--TG-----TGC-GTGTGCTC-------GTGA-TAGC--------------------------TCGAAACAGTGTGACCATACATCTTTATATGTATATAATTATATATATACATACA-------CTAATACAT------AAGCACACGCGTACATGGC-------------------------------- |
| droGri2 |
scaffold_15252:4547485-4547767 + |
|
TTGCCTTTTCC-----TGCACAGCCA----AGCAGAGCCAAGCG-----------------------------------------------------------------------------------------------------------GAGT----------CGCG-A---GCGT---GGTCGTG-CC------------------------TGTGTCTGTGTCTGTGTCTGCGACTGTCTGTGAC------------------------------TGTGAC----TGTG--------------ACTTGTGTATGTG----------CGTGCGTG----CG--------------TGCTTGTGTGTGTGGTGT-GTGTCTGTGTCTT---GGTC----TGT--GTCTCTG------------------------TGTGTG--------T------GTG----T------GTGCTTGTTGTCTGTC-GTCGCTTAACACTGCGCGCGC--GTTTTGTCATC--------------------GTGTGA--GTGGATGTATT-TGTCT------------GTGTGTGCGT---CTGTGTGTGTGTGTGCGT------GTGT--------------G----------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |