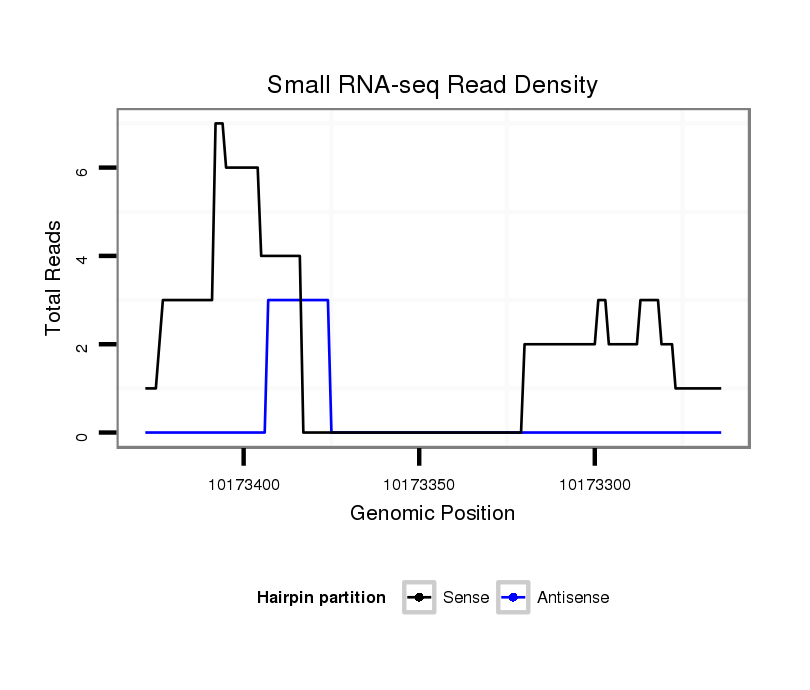
ID:dsi_18486 |
Coordinate:3r:10173314-10173378 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
intergenic
No Repeatable elements found
|
TGGTTTGCTACTACGGCTGCCGAGCATTTGGGCATTGAAAAGGGCAAATGGTAAGAGGTAGAAATCCTCCGTCAAAGGAAATCTCTAACATATGTGTTCGTTAATCCTAATGCAGTTGGTCACCTCGCAAGGCGCATCACGAGCGGATGACGCTCCTGAAGTCTG
**************************************************................((..((...((((.......(((......)))......))))...))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
M023 head |
SRR553485 Chicharo_3 day-old ovaries |
SRR902008 ovaries |
M053 female body |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................CGAGCATTTGGGCATTGAAAAGGGC........................................................................................................................ | 25 | 0 | 1 | 4.00 | 4 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................AATGCAGTTGGTCACCTCGCAAGG................................. | 24 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| TGGTTTGCTACTACGGCTGCCGA.............................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....TGCTACTACGGCTGCCGAGCATTTGGGC.................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CGCATCACGAGCGGATGAC.............. | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....TTGCTACTACGGCTGCCGAGCATTTGGGC.................................................................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................AGGCGCATCACGAGCGGA.................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........CTACGGCTGCCGAGCATTTTGGC.................................................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................AGCGGATGACGCTCCTGAAGTCTG | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TCGCAAGGCGCACCACGAGCGGATGACG............. | 28 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................GTTAGAATTCCTCCGTC............................................................................................ | 17 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................ACCTCTAACATATGTGCA................................................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................TGCTGAGCATTGGGGCATTT................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................ATTTGGGCGTTGAAACGGT......................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
ACCAAACGATGATGCCGACGGCTCGTAAACCCGTAACTTTTCCCGTTTACCATTCTCCATCTTTAGGAGGCAGTTTCCTTTAGAGATTGTATACACAAGCAATTAGGATTACGTCAACCAGTGGAGCGTTCCGCGTAGTGCTCGCCTACTGCGAGGACTTCAGAC
**************************************************................((..((...((((.......(((......)))......))))...))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
O001 Testis |
O002 Head |
SRR553488 RT_0-2 hours eggs |
M025 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................TTAGGATTTCGTCAACAAGTGGT........................................ | 23 | 3 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| ...................................ACTTTTCCCGTTTACCAT................................................................................................................ | 18 | 0 | 1 | 3.00 | 3 | 0 | 2 | 1 | 0 | 0 |
| .......................................................................................................AAGGATTTCGTCAACAAGTGGAG....................................... | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................AATCTTTAGGAGGCA............................................................................................. | 15 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................AGGAGGCAGTTTAATTTA................................................................................... | 18 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 |
| .CCAAACCATCATGCCGAT.................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 10:35 AM