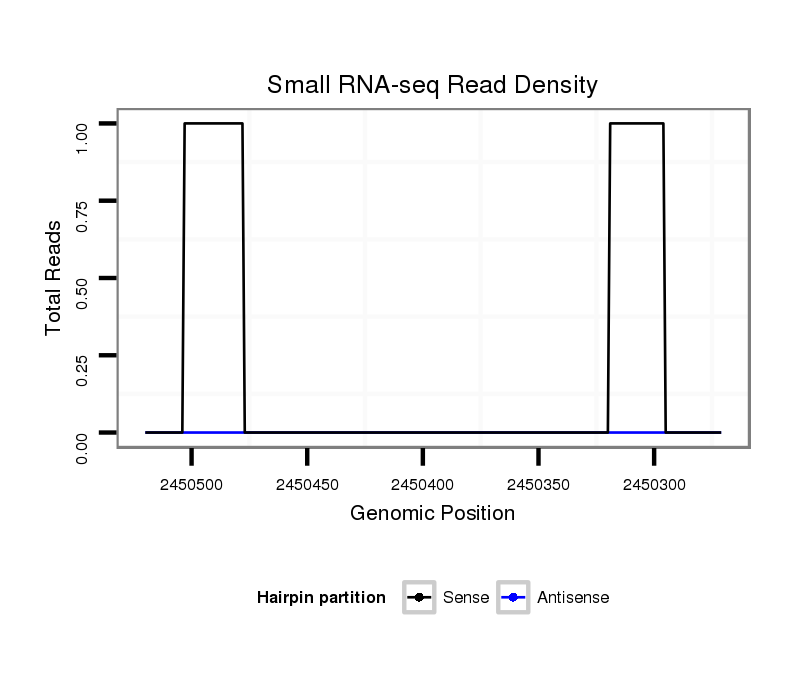
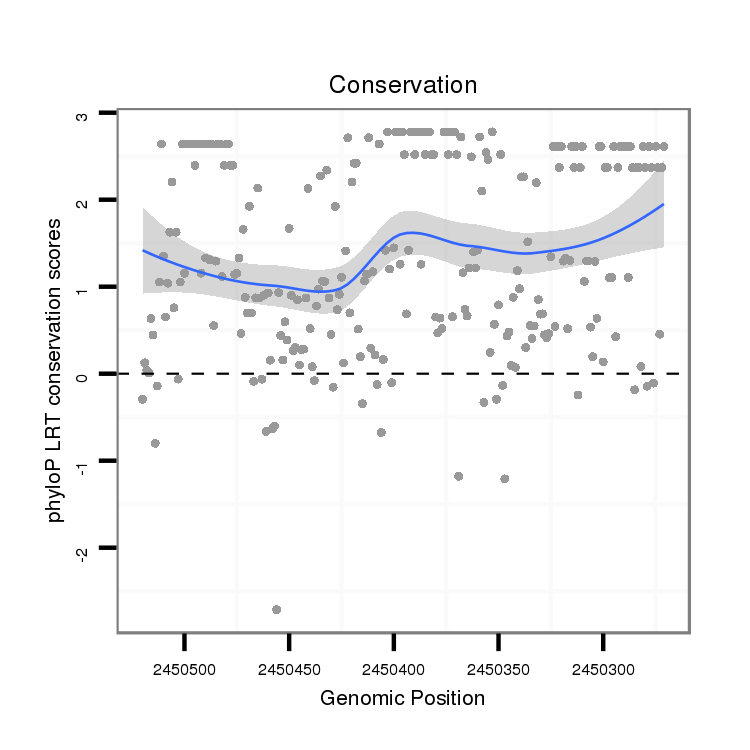
ID:dsi_1846 |
Coordinate:3l:2450321-2450470 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [3l_2449397_2450320_-]; CDS [3l_2449397_2450320_-]; intron [3l_2450321_2451250_-]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ACCTACCGGAATGAAGAGCAGAATTCATGAAAAACTTTCCATGCTGTGGTGGGTGGGGACGAACCAAGCTCTCTTTTAATCGATATTTAGATGCCTCTAAGTATTAAAAACAGACCAACCTAACACCATTCTTCTCTTGCTGCGTACTTACTACAACAACAACAACTACTACTCCTCCCTGAACCCGAAATCCCCCATAGAAACTAGAGCAGAACATAAACCGCCTGCAAAAGTCCGCATCCACATCGCA **************************************************(((((((((.((.......(((.((((....((((((((((....)))))))))).)))).)))......................(((.((.(((.....)))..)).)))..........)).)))))..))))..............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M024 male body |
SRR902008 ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
GSM343915 embryo |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................GCAGAATTCATGAAAAACTTTCCATG............................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................CATAGAAGCTAGAGCAGTA.................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................AACTAGAGCAGAACATAAACCGCC......................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................TGCTTCTCTTGCTCGGTACTT..................................................................................................... | 21 | 3 | 3 | 0.67 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ................................AACTGTCCATGCTGT........................................................................................................................................................................................................... | 15 | 1 | 6 | 0.50 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| ..............................................................................AGCGACATTGAGATGCCTCTA....................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................TGCCTCTAAGTATTATAGCC........................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................GTACTTACTACAACAACGGAAA..................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................................CAACAACAACTCGTAGTCCTC......................................................................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................ACAACTGTCCATGCTG............................................................................................................................................................................................................ | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
TGGATGGCCTTACTTCTCGTCTTAAGTACTTTTTGAAAGGTACGACACCACCCACCCCTGCTTGGTTCGAGAGAAAATTAGCTATAAATCTACGGAGATTCATAATTTTTGTCTGGTTGGATTGTGGTAAGAAGAGAACGACGCATGAATGATGTTGTTGTTGTTGATGATGAGGAGGGACTTGGGCTTTAGGGGGTATCTTTGATCTCGTCTTGTATTTGGCGGACGTTTTCAGGCGTAGGTGTAGCGT
**************************************************(((((((((.((.......(((.((((....((((((((((....)))))))))).)))).)))......................(((.((.(((.....)))..)).)))..........)).)))))..))))..............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M053 female body |
M024 male body |
SRR618934 dsim w501 ovaries |
SRR1275487 Male larvae |
M023 head |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
SRR1275483 Male prepupae |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR1275485 Male prepupae |
M025 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................ATGAGGGGGGTCTTGGGTT.............................................................. | 19 | 3 | 20 | 1508.80 | 30176 | 29399 | 495 | 65 | 78 | 42 | 17 | 16 | 30 | 6 | 16 | 5 | 3 | 3 | 1 |
| .........................................................................................................................................................................ATGAGGGGGGTCTTGGGTTT............................................................. | 20 | 3 | 12 | 665.33 | 7984 | 7720 | 163 | 14 | 31 | 14 | 12 | 9 | 8 | 1 | 5 | 1 | 4 | 2 | 0 |
| ..........................................................................................................................................................................TGAGGGGGGTCTTGGGCT.............................................................. | 18 | 2 | 2 | 30.00 | 60 | 57 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TGAGGGGGGTCTTGGGCTT............................................................. | 19 | 2 | 1 | 16.00 | 16 | 15 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGTCTTGGGCT.............................................................. | 19 | 2 | 1 | 15.00 | 15 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TGAGGAGGGTCTTGGGTT.............................................................. | 18 | 2 | 6 | 9.00 | 54 | 54 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGTCTTGGGTTTT............................................................ | 21 | 3 | 3 | 8.67 | 26 | 20 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGAGGGTCTTGGGTT.............................................................. | 19 | 2 | 2 | 6.50 | 13 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TGAGGAGGGTCTTGGGTTT............................................................. | 19 | 2 | 3 | 6.00 | 18 | 17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................GATGAGGGGGGTCTTGGGTTT............................................................. | 21 | 3 | 4 | 5.50 | 22 | 21 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................TATGAGGGGGGTCTTGGGCTT............................................................. | 21 | 3 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................GAGGGGGGTCTTGGGCTT............................................................. | 18 | 2 | 4 | 4.25 | 17 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGAGGGTCTTGGGTTT............................................................. | 20 | 2 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGTCTTGGGCTT............................................................. | 20 | 2 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................GATGAGGGGGGTCTTGGGTT.............................................................. | 20 | 3 | 8 | 3.75 | 30 | 28 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGCCTTGGGTTT............................................................. | 20 | 3 | 6 | 2.67 | 16 | 7 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 1 | 0 | 2 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGACTTGGGTT.............................................................. | 19 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGACTTGGGTTT............................................................. | 20 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGTCTTGGGTTTTA........................................................... | 22 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TGAGGGGGGACTTGGGTT.............................................................. | 18 | 2 | 3 | 2.00 | 6 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGTCTTGGGGT.............................................................. | 19 | 3 | 20 | 1.80 | 36 | 34 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................TATGAGGAGGGTCTTGGGTT.............................................................. | 20 | 3 | 6 | 1.33 | 8 | 7 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TGAGGGGGGTCTTGGGCTTC............................................................ | 20 | 3 | 9 | 1.11 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TGAGGGGGGTCTTGGGCTTT............................................................ | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TGCTAAGAAGAGAACGCCGTAT........................................................................................................ | 22 | 3 | 2 | 1.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................GCACGACGGATGAAGGATGTT.............................................................................................. | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGTCTTGGGCTTC............................................................ | 21 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGTCTTGGGC............................................................... | 18 | 2 | 3 | 1.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................TTTGGCGGGCGTTTTCAGGCG............ | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................GAGGAGGGTCTTGGGTTT............................................................. | 18 | 2 | 12 | 0.67 | 8 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TGAGGGGGGTCTTGGGTTTTA........................................................... | 21 | 3 | 5 | 0.60 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGGCTTGGGTTT............................................................. | 20 | 3 | 11 | 0.55 | 6 | 4 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................TATGAGGGGGGTCTTGGGCT.............................................................. | 20 | 3 | 4 | 0.50 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGCCTTGGG................................................................ | 17 | 2 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................GATGAGGCGGGTCTTGGGTTT............................................................. | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGAGGGTCTTGGGTTTC............................................................ | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TGAGGGGGGACTTGGGTTT............................................................. | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGCCTTGGGCT.............................................................. | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGCCTTGG................................................................. | 16 | 2 | 15 | 0.47 | 7 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 3 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGTCTTGGG................................................................ | 17 | 2 | 8 | 0.38 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TGAGGAGGGTCTTGGGGTT............................................................. | 19 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGCGGGTCTTGGGTTT............................................................. | 20 | 3 | 6 | 0.33 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGAGGGTCTTGGGT............................................................... | 18 | 2 | 6 | 0.33 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................GAGGGGGGACTTGGGTTT............................................................. | 18 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................TCTATCAGTCTACGGAGATT...................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................TATGAGGGGGGACTTGGGTT.............................................................. | 20 | 3 | 8 | 0.25 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................GATGAGGGGGGCCTTGGGTT.............................................................. | 20 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................TGTTGTTGTTGTTGATGATGA............................................................................. | 21 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGGTCTTGGGGTT............................................................. | 20 | 3 | 8 | 0.25 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................CGATTTCAGGCGTAGGCGAAG... | 21 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................TGGTGAGGATGAGGAGGGA...................................................................... | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................TGAGGAGGGCCTTGGGGT.............................................................. | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................ACCACGGCTTGGTTCGCGAG................................................................................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................TATGAGGGGGGTCTTGGGC............................................................... | 19 | 3 | 12 | 0.17 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................TGTTGTTGTTGTTGATGATG.............................................................................. | 20 | 0 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................AGGGGGGTCTTGGGCTT............................................................. | 17 | 2 | 15 | 0.13 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATGAGGGGGCTCTTGGGCT.............................................................. | 19 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................GATGAGGGGGGTCTTGGGT............................................................... | 19 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................GAGGGGGGTCTTGGGCT.............................................................. | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................AGAACGACGCAAGAA..................................................................................................... | 15 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................TGAGGAGGGTCTTGGGTTTG............................................................ | 20 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................ACGACGCAAGAACGATG................................................................................................ | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................GGAGTTATCTTTGATCGC......................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3l:2450271-2450520 - | dsi_1846 | A-CCTACCGGAATG--AA-GAGCAGAATTCATGAAAAAC-TTTCCATGCTGTG---------------------------GTGGGTGGGGACGAAC------------------------------------------------------C-AAGCTCTCTTTTAA--------------------------------------TCGAT--ATTTAGA-T--GCC----------T--CT-AAGTA------TTAA---------------------------------------------------------------------------------------------AAACAGACC---AACCTAACA-C---CATTCTTCTCTTGC----TGCG-TACTTACTA-CAACA---------------------ACAACAAC-------------------------TA---C------------T-------------------------------ACTCCTCCCTGAA--C-------C-----------CGAA-----------------------------------------------------------ATCCCC-------------------CATAGAAACTAGAGCAGAA---CATAAACCGCCTGCAAAAGTCCGCATCCACATCGCA-- |
| droSec2 | scaffold_2:2583412-2583661 - | dse_1001 | A-CCTACCGGAATG--AA-GAGCAGAATTCATGAAAAAC-TTTCCATGCTGTG---------------------------GTGGGTGGGGACGAAC------------------------------------------------------C-AAGCTCTCTTTTAA--------------------------------------TCGAT--ATTTAGA-T--GCC----------T--CT-AAGTA------TTAA---------------------------------------------------------------------------------------------AAACAGACC---AACCTAACA-C---CATTCTTCTCTTGC----TGCG-TACTTACTA-CAACA---------------------ACAACAAC-------------------------TA---C------------T-------------------------------ACTCCTCCCTGAA--C-------C-----------CGAA-----------------------------------------------------------ATCCCC-------------------AATAGAAACTAGAGCAGAA---CATAAACCGCCTGCAAAAGTCCGCATCCACATCGCA-- |
| dm3 | chr3L:2562794-2563057 - | A-CCTACCGGAATG--AA-GAGCAGAATTCATGAAAAAC-TTTCCATGCTGTG---------------------------GTGGGTGGGGACGAAA------------------------------------------------------CAAAGCTCTCTTTTAA--------------------------------------TCGAT--ATTTAGA-T--GCC----------T--CT-AAGTA------TTAAAAAAAAAA---------------------------------------------------------------------------------AAAAAAACAGACC---AACCTAACA-C---CATTCTTCTCTTGC----TGCG-TACTTACTA-CAACA---------------------ACAACTAC-------------------------TA---C------------T-------------------------------CCTCCTACCTGAA--C-------C-----------CGAA-----------------------------------------------------------ATCCCC------------------AAATAGAAACTAGAGCAGAA---CATAAACCGCCTGCAAAAGTCCGCATCCACATCGCA-- | |
| droEre2 | scaffold_4784:2578987-2579240 - | A-CCTACCGGAATT--AA-GAGCAGAATTCATGAAAAAC-TTTCCATGCTGTG---------------------------GTGGGTGGGGACGAAC------------------------------------------------------C-AAGCTCTCTTTTAA--------------------------------------TCGAT--ATTTAGA-T--GCC----------T--CT-AAGTA------TTAAA--------------------------------------------------------------------------------------------AAACAGACC---AACCTAACA-C---CATTCTTCTCTTGC----TGCG-TACTTACTA-CAACA---------------------ACAACAACT-------------------A---CTA---C------------T-------------------------------ACTCCTCCCTGAA--C-------C-----------TGAA-----------------------------------------------------------ATCCCC-------------------AATAGAAACTAGAGCAGAA---CATAAACCGCCTGCAAAAGTCCGCCTCCACATCGCA-- | |
| droYak3 | 3L:9376620-9376878 + | A-CCTACCGGAATG--AA-GAGCAGAATTCATGAAAAAC-TTTCCATGCTGTG---------------------------GTGGGTGGGGATGAAG------------------------------------------------------C-AAGCTCTCTTTTAA--------------------------------------TCGAT--ATTTAGA-T--GCC----------T--CT-AAGTA------TTAA---------------------------------------------------------------------------------------------AAACAGACCAACAACCTAACA-C---CATTCTTCTCTTGC----TGCG-TACTTACTA-CAACA---------------------ACAACCAC-------------------------TA---C------------T-------------------------------ACTCCTCCCTGAA--C-------C-----------TGAA---------CCTGAA--------------------------------------------ATCCCC-------------------AATAGAAACTAGAGCAGAA---CATAAACCGCCTGCAAAAGTCCGCCTCCACATCGCA-- | |
| droEug1 | scf7180000409466:288110-288389 - | A-CCGACTGGAATG--AA-GAACAGAATTCATGAAAAAC-TTTCCATGCTGTG---------------------------GTGGGTGGGGAA------------------------------------------------------AGAAGAAAGCTCTCTTTTAA--------------------------------------TCGAT--ATTTAGA-A--GCC----------T--CT-AAGTA------TTAAA--------------------------------------------------------------------------------------------AAACAGAAC---AACCTAACA-C---CATTCTTCTCTTGC----TGCG-TACTCACCA-CAACAACAA---------CAACAACAACTACTAC-------------------------TA---C------------T-------------------------------ACTACTCCCTGAA--C-------C-----------TGAA---------CCTGAACCTG-------------------------------ATA------TTCCCC----------------AAAAATAGAATCTAGACCAGAA---CATAAACCGCCTGCAAAAGTCCGCCTCCACATCGCA-- | |
| droBia1 | scf7180000302428:9332341-9332585 - | AGTCTACCAGAATG--AA-GAGCAGAATTCATGAAAAAC-TTTCCATGCTGTG---------------------------GTGGGTGGGGACGATC------------------------------------------------------G-AGGCTCTCCT----------------------------------------------------------------------------TT-AAGTA------CTCAAG------------------------------------------------------------T------A----------------------AAACCAGACC---AACCTAACA-C---CATTCTTCTCTTGC----TGCG-TACTCACCA-CAACA---------------------ACAA---C-------------------------TA---C------------T-------------------------------ACTCCTCCCTGAA--C-------C-----------TGAA--------ACCCGAAAATG-----------------------------------------TCCCC----------------CCGAATAGAAACTAGAGCAGAA---CATAAACCGCCTGCAAAAGTCCGCCTCCACATCGCA-- | |
| droTak1 | scf7180000415857:335272-335575 - | ---CTACTGGAATGTGAA-GAGCAGAATTCATGAAAAAC-TTTCCATGCTGTG----------------TTT-------GGTGGGTGGGGACGAAC----------------CTCCAAAAGGTC--------------------------T----CTCTCTTTTAA--------------------------------------TCGTTAAATTTAGA-A--GCCTCCT----CCT--CC-AAGTATATATATTAA---------------------------------------------------------------------------------------------AAACAGACC---AACTTAACACC---AATTCTTCTCTTGC----TGCG-TACTCACAA-CTACA---------------------ACAACAAC-------------------------AA---C------------T-------------------------------ACTACTCCCTGAA--C-------C-----------TGAA-------------------------------AAATCCAA--CAAAACCCAAAACAAAAAA-------------------AAACAAATAGAAACTAGAACAGAA---CATAAATCGCCTGCAAAAGTCCGCCTCCACATCGCA-- | |
| droEle1 | scf7180000491249:2839067-2839316 + | A-CATTCCGGAATG--AA-GATCAGAATTCATGAAAAAC-TTTCCATGCTGTG---------------------------GTGGGTGGGAAT-C-----------------------------------------------GAAA-AAAAGAAAACA--------------------------------------------------------------------TACTCTATACT--CT-AAGTA------TTAAAAAAA----------------------------------------------------------------------------------ACCAACAAACAGACC---AACTTAACA-C---CATTCTTCTCTTGC----TGCG-TACTCACA--------------------------------------------------------------A---C------------T-------------------------------ACTCCTCCCTGAAAAC-------C-----------TG------------CTGAA----------------AA--------------------------ATCACT-------------AAAAAAAATAGAAACTAGAGCAGAA---TATAAACCGCCTGCAAAAGTCTGCCTCCACATCGCA-- | |
| droRho1 | scf7180000779552:7333-7596 + | A-CCTACTGGAATG--AA-GAGCAGAATTCATGAAAAAC-TTTCCATGCTGTG---------------------------GTGGGTGGGGACGAAC------------------------------------------------------C-CAGATCTCTTTTAA--------------------------------------TCAAT--ATTTAGA-A--GCC----------T--CT-AAGTA------TTAAACAAAAAA-------------------------------------------------------------------------------AACAAAAAACAGACC---AACTTAACA-C---CATTCTTCTCTTGC----TGCG-TACTCACTA-CAACA---------------------ACAA---C-------------------------TA---C------------T-------------------------------ACTCCTCCCTGAA--C-------C-----------TGCA-----------------------------------------------------------ATCCCC----------------CCAAATAGAAACTAGAGCAGAA---CATAAACCGCCTGCAAAAGTCCGCCTCCACATCGCA-- | |
| droFic1 | scf7180000454105:886060-886371 - | AACCTGCTGGAGCG--AA-GAGCAGAATTCATGAAAAAC-TTTCCATGCTGTG---------------------------GTGGGTGGGG-CGATC------------------------------------------------------C-AAGCTCTCT-TTAA--------------------------------------TCGAT--ATTTAGA-A--GCC----------T--CT-AAGTA------TTCAAAAG----------------------------------------------------------C------A------------------------AACAAAAG-CAAACTTAACA-C---CATTCTTCTCTTGC----TGCG-TACTCACCA-CAACA---------------------ACAACAACA-------------------A---CTA---C-------CCAACA-------------------------------AC--TTCCCCGAA--T-------C-----------TGAA---------TCTGAATCTGAATCTGAATCTGAAATCGAAACCGAAACCCAACCGAATCAATCCCC-------------------AATAGAAACTAGAGCAGAA---CATAAACCGCCTGCAAAAGTCGGCCTCGACCTCGCA-- | |
| droKik1 | scf7180000302441:1139152-1139409 - | A-TCTAAT----------------------------------------------------------------------------------------------------------------------------------------------------TCTTTGTTGA--------------------------------------TTAGTGTATTTAGTAT--GCC----------TCTCT-AAGTA------TGCAA-------------------------------------------------------------------------------------------AAATCCGATCA-AAACTTAACA-C---AATTCTTCTCTTGC----TGCG-TACTCACAAACAACAACAA---------CTACAACTACAACAAC-------------------------AA---C------------T-------------------------------ACTCCTCCCTGAAAACAATCAAACAAAAAAATCCCTGAAATCCTGAAAAATGAA------------------------------AC-CAATCGAA------CAC----------------ACAAATAGAAACTAGAGCAGAACAACATAAATCGCCTGCAAAAGTCCGCCTCGACTTCGCA-- | |
| droAna3 | scaffold_13337:1778319-1778662 + | dan_2346 | G-C-AACGCGAATG--------CAGAATTCATGAAAGAC-TTTCCATGCTGTG----------------T---------GGTGGGAGGGGAT-CCTAGTCTAAAGTCCTAGAGTCCTAGAGGCC--------------------------T-AGGCTCTT-TCTAA--------------------------------------T-GAT--ATCTAGAAAACGCC----------T--CT-AAGTA------TGCA---------------------------------------------------------------------------------------------------ACC---GACTTAACA-C---ACTTCTTCTCTTGC----TGCG-TACTCACCAACAACAACAA---------CAACA---ACAACAAC----------------------AACTACAACCA--------------------------AACATCTACAACAACTACTACTCCCTGAA--T-------CCAAAAAATATGTTAAATCCTGAATCCTGAA--------------------------------------------ACCCCC--TGACAAAAAAAAAAAAAAACAGAAACAAGAACAGAA---CGCCAATCGCCTTCAAAAGTCCGCCTCCACATCCCA-- |
| droBip1 | scf7180000396544:312453-312698 - | T-------TGAATG--------CAGAATTCATGAAAGAC-TTTCCATGCTGTG----------------T---------GGTGGGCGGGG----------------------------------------------------------------------TTCTAG--------------------------------------TC-------CTAGA-T--GAT----------A--C--------------------------------------------------------------------------------------------------------------CAACCGACC---AACTTAACA-C---AATTCTTCTCTTGC----TGCG-TACTCACCAACAACAACAA---------CA------ACAACAACT-------------------ACTACTACAAACAACCAACCAACA-------------------------------ACCCCCCCTTGAA------------------------AA-----------------------------------------------------------ACCCCCCTTGAT--------AAAAAAATAGAAACAAGAACAGAA---CGCGAATCGACTTCAAAAGTCCGCCTCGACATCCCA-- | |
| dp5 | XR_group3a:1368160-1368390 - | T-C----GCGAAG---------GAGAATTCATGAAAAAT-GTTACATGCTGTT---------------------------GTGGTGGGGCAT-A--------------------------------------------------------G-AA--TCTCCTCCTACTCCTACCCCTCCCCACTCCCACCCCCACCCCCACCCTTCGTC--CTTTAGAAT-CGCC----------T--CT-AAGTA------TGCAACA------------------------GAGACTGCAAGAACAA-AAACAACAACCAACAAAGT------A----------------------AAAAAACAAA---AACCTAACA-CACAAATTCTTCTCTTGC----GCTGCTACTCACAA-CAACA---------------------ACAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_5:821604-821634 - | C-C-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A-CAGCA---------------------ACAACAAC-------------------------TA---C------------T-------------------------------ACGGTTCCCTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004729:2190420-2190770 + | A-CAAAAAGGAATGC-AA-GAGGAAAATTCATAAGAAATGTTTCCATGCTGTTGCAGCAATCTACTATTTTTT-TTCT---ATGGTGGGGA--------------------------------------------------------------------------------------------------------------------AT--ATTAATT-T--GCC----------TAGCTAAAGTA------ATGCAACA-----------------------GACAAAAAAAAAAAAAGACAAAATAATTAACACAAA------A----------------------AAATCAAAAC---AAAAAAATA-C-CAAATTCTTTTCTTGCGCGTTGCC-TACTTACCA-AACT----------------------GTAA-AACTGCCCCGAACTCCCCCTTTACCACCTAC-CACCACCAACCAACACCTCTGACCCTACCCAACCTCTATTCCAACTGC----------------------------------------------------------------------------------------------------------------------GACAACAGATACTAGATCAGGG---TATAAATCGCTTGCAAAAATCAGCCTCGACATCACA-- | |
| droVir3 | scaffold_13049:5183091-5183349 - | dvi_18411 | G-G--AATGCAATGCA-ATG-AGAGAATTCATGAAAAAC-TTTCCATGCGGTT----------------TTTT-TTCCA---------------------------------------------TA-CCTGGGTGCAGATAGCCA--------AA-----------------------------------------------------------------------------------ATAAAGTA------TTAAAAAAAAAAACAACAATTTAGCCTGTAG------------------------------------AACTGTAAAATGCAAGC-------------AAACA-AAA---AACCTAACAAC---AATTCTTCTCTTGC----GCTACTACTCACAA-CAACA---------------------ACAACGAC-------------------------AA---C------------A-------------------------------ACT---------------------------------------------------------------------------------------------------------------------GTAAACAGAAATTAGAACAAAA---CATAAACCGCTTACAAAAATCGGCATCCACATCGCA-- |
| droMoj3 | scaffold_6680:3553619-3553891 - | dmo_2332 | G-G--AATGCAATGCA-ATG-AGAGAATTCATGAAAAAC-TTTCCATGCGGTT----------------TTTTTTTTCA---------------------------------------------TA-CCTGGGTGTAGATAGCCAAAAAAAAAAACA-ACTTAGAA--------------------------------------T---------------------------------GT-AG----------------------------------------------------------------------------AAATGTAAAATGCAAGTGAACAAAACCA--AAACAAACA-AAAACCTAACAAC---AATTCTTCTCTTGC----GCTGATACTCACAA-CAACA---------------------ACAACAAC-------------------------AA---C------------C-------------------------------ACAACATGTTAAA--C----------------------A-----------------------------------------------------------ACT-------------------GTAAACAGAAACTAGAACAAAA---CGTAAATCGCTTACAAAAATCCGCTTCCACTTCGCA-- |
| droGri2 | scaffold_15110:9801536-9801806 + | G-G--AATGCAATGCA-ATC-AGAGAATTCATGAAAAAC-TTTCCATGCTACA--AGCGGTT-------TTTT-TT----------TGTTGT-A------------------------------TACCCTGGGC----------AAGAAAC-AA---------------------------------------------------------ATTTAGA------C----------T--GT-AG----------------------------------------------------------------------------AAATGTA--ATGC------------------AACCG--A---AACCTAACAAC---AATTCTTCTCTTGC----GCTGTTACTCACAA-CTACAACAATTATTTGTACAACAACAACAACGAC-------------------------AA---C------------C-------------------------------ACATCTACATGAA--C----------------------A-----------------------------------------------------------ACT-------------------GTAAACAGAAATTAGAACAAAA---CGTAAATCGCTTACAAAAATCCGCTTCCACCTCGCATG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/19/2015 at 05:42 AM