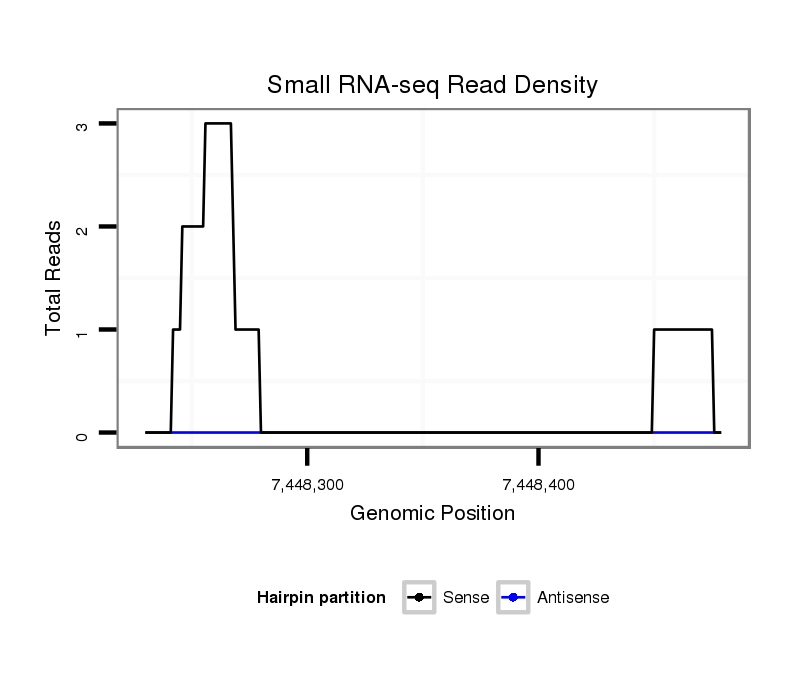
ID:dsi_1837 |
Coordinate:3l:7448280-7448429 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
intergenic
No Repeatable elements found
|
GATCTCGAAGCCAGGAACCAAAGGAATCGCGAGCTCGTTAGCGTTCCAGGGTAAGCCTGCGTCTGGCATCGTAAACAAATGCCCGTCAAAAGGCATTCAGCTAAACACCGACAAAGTAGCAAACAAAATATATGTGGACAGCCAGCCCTCCATTTTGTATATGTGTATAATATGGGAACTTGCCAAGTTCAAGGCAGGAGTTGAAGATGTCTATATATGATGCATTGTATCCACAATTATGGCAGAGGGA
**************************************************.....(((((..(((((...((..(((((((((........))))))..((((.............))))............)))..)).)))))....((((.((((((....)))))).))))((((((...))))))...)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
GSM343915 embryo |
SRR902009 testis |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................................TGCATTGTATCCACAATTATGGCAGA.... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................ACCAAAGGAATCGCGAGCTCGT.................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................TCGCGAGCTCGTTAGCGTTCCAGG........................................................................................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............AGGAACCAAAGGAATCGCGAGCTCGTT................................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................AAGGAATCGAGAGCTC...................................................................................................................................................................................................................... | 16 | 1 | 4 | 0.75 | 3 | 0 | 1 | 2 | 0 | 0 | 0 | 0 |
| .....................AGGAATCGAGAGCTC...................................................................................................................................................................................................................... | 15 | 1 | 9 | 0.56 | 5 | 2 | 1 | 2 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................TCAAAGCAGGACTTGAAGAGGT........................................ | 22 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................TGTGGACAGCGATCGCTCCA.................................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................ATTTAGTATACGGGTATAATA.............................................................................. | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................AAGGAATCGAGAGCT....................................................................................................................................................................................................................... | 15 | 1 | 11 | 0.18 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..................CCAAGGAATCGAGAGCTC...................................................................................................................................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................GTGGACAGCCAGCTC...................................................................................................... | 15 | 1 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................................................................TTCAAGCCAGGAGCTGA.............................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................ATCGCAAGATCGTTAGC................................................................................................................................................................................................................ | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CTAGAGCTTCGGTCCTTGGTTTCCTTAGCGCTCGAGCAATCGCAAGGTCCCATTCGGACGCAGACCGTAGCATTTGTTTACGGGCAGTTTTCCGTAAGTCGATTTGTGGCTGTTTCATCGTTTGTTTTATATACACCTGTCGGTCGGGAGGTAAAACATATACACATATTATACCCTTGAACGGTTCAAGTTCCGTCCTCAACTTCTACAGATATATACTACGTAACATAGGTGTTAATACCGTCTCCCT
**************************************************.....(((((..(((((...((..(((((((((........))))))..((((.............))))............)))..)).)))))....((((.((((((....)))))).))))((((((...))))))...)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................GAGTTGTCGCTGCTTCATCGT................................................................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................................TCAATTTGTGGATGTTTCAT.................................................................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................CGTTTGTTTCCGGGCAGTTT................................................................................................................................................................ | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................AGATCTATACTACGTCA........................ | 17 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 |
| ..............................................................................TACGCGCAGTAGTCCGTAA......................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 09:32 AM