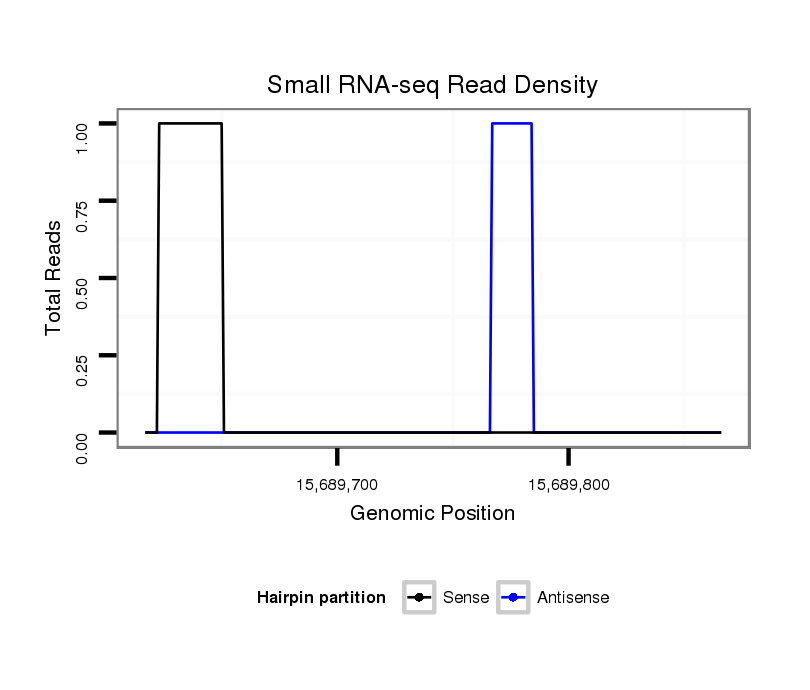
ID:dsi_18297 |
Coordinate:3l:15689667-15689816 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [3l_15689580_15689666_+]; CDS [3l_15689580_15689666_+]; intron [3l_15689667_15692625_+]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AAAGAAGAGCTATGGAACGCAAGCTTTCCGAGTTGGAAGAAGAGCTCAAGGTAATTACAAAAGTGTCCTTTGCTAACCGTAAGCGTTAAAAGTGCCGCCGCTGTTGCTGTTTTTGTGTTCTGAGAACAGGACCCCCACATTGATTGCTATATTGGTTTATTTTTATTTTGATTCCATTCCCCACATGATTTGTCCAAGAGTTTGTGATACTTTTCCCCCAGAAAAGCCTTGTTGCATGTCTCAAATTCTG **************************************************....((((((..((((.............(((....((((((((..((((.(((.((.(((..(((((((((....))))))....)))..))).)).))).)))).))))))))..)))............))))...)))).....))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
SRR553488 RT_0-2 hours eggs |
M025 embryo |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|
| ......GAGCTATGGAACGCAAGCTTTCCGAGTT........................................................................................................................................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................GTTGATTGCTATAATGGT.............................................................................................. | 18 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................TTTGATTCCATTCCCCC................................................................... | 17 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................TACCCCACATGATTCGGCC....................................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................................................AGTGCTGCCGCTGTTG................................................................................................................................................ | 16 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
|
TTTCTTCTCGATACCTTGCGTTCGAAAGGCTCAACCTTCTTCTCGAGTTCCATTAATGTTTTCACAGGAAACGATTGGCATTCGCAATTTTCACGGCGGCGACAACGACAAAAACACAAGACTCTTGTCCTGGGGGTGTAACTAACGATATAACCAAATAAAAATAAAACTAAGGTAAGGGGTGTACTAAACAGGTTCTCAAACACTATGAAAAGGGGGTCTTTTCGGAACAACGTACAGAGTTTAAGAC
**************************************************....((((((..((((.............(((....((((((((..((((.(((.((.(((..(((((((((....))))))....)))..))).)).))).)))).))))))))..)))............))))...)))).....))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O002 Head |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
M024 male body |
SRR553486 Makindu_3 day-old ovaries |
M023 head |
M053 female body |
SRR553485 Chicharo_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................TAACCAAATAAAAATAAA.................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................GCGGAGACAACGACAAATACA...................................................................................................................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................TCTTATCCAGTTCCATTAA.................................................................................................................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................TCTTATCCAGTTCCAGTAA.................................................................................................................................................................................................. | 19 | 3 | 20 | 0.50 | 10 | 0 | 0 | 0 | 5 | 0 | 0 | 4 | 0 | 1 | 0 |
| ............................................................................................ACGGCGGCGACAACG............................................................................................................................................... | 15 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............CTTGAGTTCCAAAGGCGCAA........................................................................................................................................................................................................................ | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................TCTTCGCGAGTTCCAT..................................................................................................................................................................................................... | 16 | 1 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................GCAATGCTCAGGGCGGCG..................................................................................................................................................... | 18 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| .......TCGATAGCTTCCGTTCG.................................................................................................................................................................................................................................. | 17 | 2 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................GAAATAAAAAGAAAATTAAGGT.......................................................................... | 22 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................ATAAAACTAAGGAAAG....................................................................... | 16 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Generated: 04/23/2015 at 06:07 PM