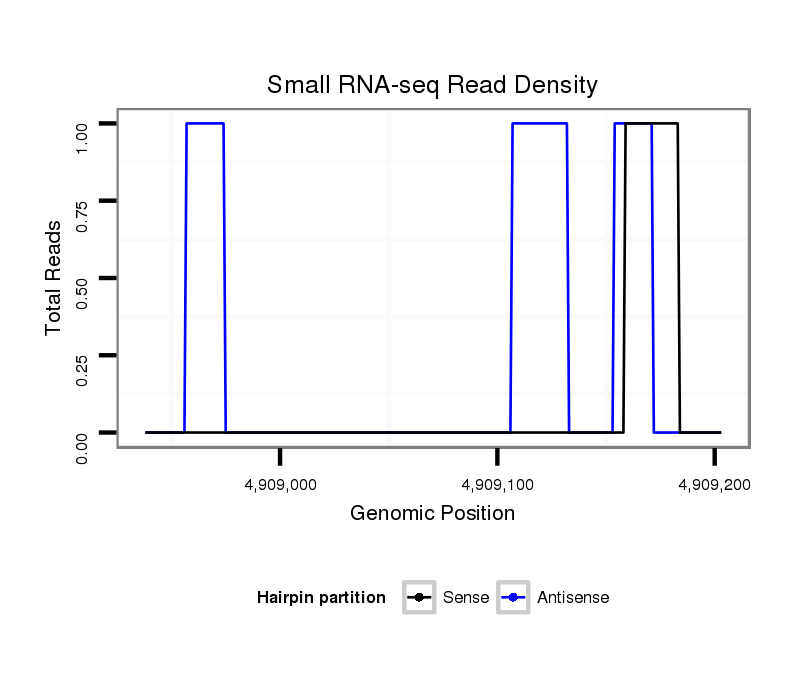
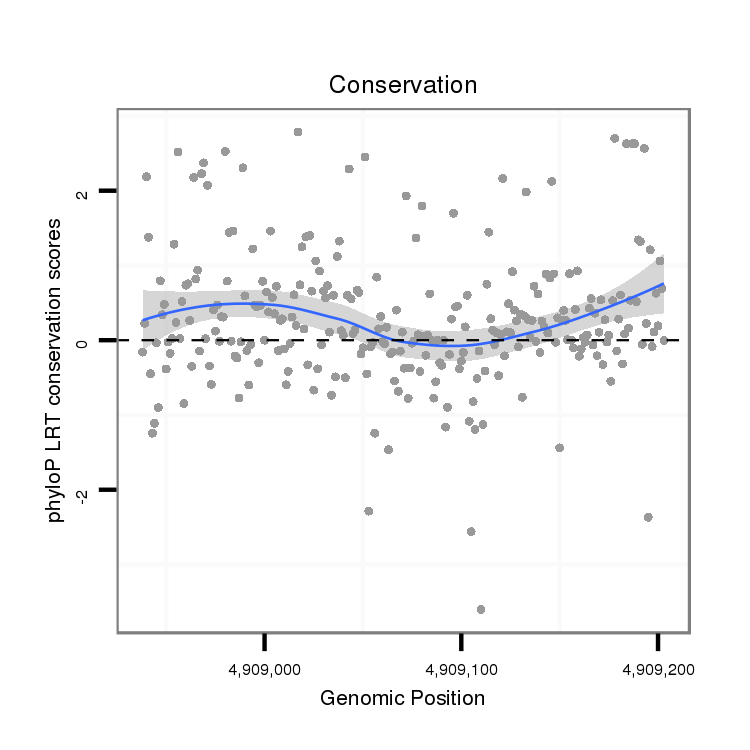
| droSim2 |
x:4908938-4909203 + |
dsi_18292 |
GA-CAGATGAAACGAAGAACAATAACTACAGCAACGGCCACAACAA---CAAAGGCATG----AACAG---CAGCA------------ACAATAA-TTTTAATAA------------CGATAA-------TAGCCACAAT--AGCAAC-----------------------------------------------TACAACAATTACGGT---TACAACAGCA------------------G-------------------------------------------------------------------------------------------------------CAGCTA------TCGTTACAGTTACCGTTACCG----TTACC------GTTGCC--------------------------------CTTACAC---T---TACA--CTTACAATTACGTTCA--------------CGAGTCCCGTT-----TTTTTTTCAG-TGCGTG------CG--TGTATGTGAAA-------------------TTGTTATTATTAACAACAACAAAAACAACAT---------- |
| droSec2 |
scaffold_33:478046-478220 - |
|
AA-CCACAACTACAAC-AACCACAA---------------CCACTA---CAACAACTTG----TACCC---CAACG------------ACTACGA-CAACCACAA------------CGACTA-------CAACCACTAC--AACGAC-----------------------------------------------TACAACTTGTACCAC---AACAACCACG------------------A-------------------------------------------------------------------------------------------------------CTACCA------CAACTACAACTACAACAACCA----CTACC------ACTACA--------------------------------ACAACTT---G---TACC--CCAACAACTACG------------------------------------------------------------------------------------------------------------------------------------ |
| dm3 |
chrX:5259431-5259671 + |
|
GA-CAGATGAAACGAAGAACAATAACTACAGCAACGGCCATAACAA---CAAAGGCATG----AACAG---CAGCA------------ACAATAA-TTTTAATAA------------CGATAA-------TAGCCACAAT--AGCAAC-----------------------------------------------TACAACAATTACGGT---TACAACAGCA------------------G-------------------------------------------------------------------------------------------------------CAGCTA------TCGTTACAGTTACC----------------------------------------------------------------------T---TACA--ATTACAATTACGTTCA--------------CGAGTCCCGTT-----TTTTTTCCAG-TGCGTG------CG--TGTATGTGAAA-------------------TTGTTAATATTAACAACAACAAAAACAACAT---------- |
| droEre2 |
scaffold_4690:2617008-2617272 + |
|
GA-CAGATGAAACGAAGAACAA-GACAACAGCAACGGCCACAACAA---CAAAGGCATG----AACAG---CAGCA------------ACAATAA-TTTTAATAA------------CGATAA-------TAGCCACAAT--AGCAAC-----------------------------------------------AGCAACAATTACAGT---TACAGCAACA------------------G-------------------------------------------------------------------------------------------------------CAGCTA------TCGTTACAGTTACAGTTACAG----TTACC------GTTACC--------------------------------GTTGCCG---T---TACA--CTTACAATTACGTTCA--------------CGAGTCCCGTT-----TTTTTTCCAG-TGCGTG------CG--TGTATGTGAAA-------------------TTGTTATTATTAACAACAACAAAACCTACAT---------- |
| droYak3 |
X:3206860-3207134 - |
|
GA-CAAATGAAACGAAGAACAAAAACTACAGCAACGGCCACAACAA---CAAAGGCATG----AACAG---CAGCA------------ACAATAA-TTTTAATAA------------CGATAA-------TAGCCACAAT--AGCAAC-----------------------------------------------AACAACAATTACAGTTACA--GTAAATTACAGTTACAGTTA----------------------------------------------------------------------------------------------C---AGCAAC---AGCAGCTA------TCGTTACAGTTACC----------------------GTTACT--------------------------------GTTGCCG---T---TACA--CTTGCAATTACGTTTA--------------CGAGTCCCGTT-----TT-TTTTCAG-TGCGTG------CG--TGTATGTGAAA-------------------TTGTTATTATTAACAACAAAAAAAACAACAT---------- |
| droEug1 |
scf7180000409182:185904-186176 + |
|
AA--AGCAAGAG-------C---AACAACGGCAACAGAAACAGCAA---CAAAGGCATG----AACAG---CAGTA------------ACAATAA-TTTTAATAA------------CGATAATAGCCAT-----ATAAT--AGCAAC---AAGAACAACAACAACAACAACAGCAACGGCAAAAACAACAA-T-TA---CAATT------------------------ACGATTA----------------------------------------------------------------------------------------------C---GATTGCAACGACGGTTA------TCGTTACAGTTACC------------------------------------------------------------------G---T---TACACTCTTACAATTACGTTGA--------------CGAGTCCCGTTATATTTTTTTTCCAG-TGCGTG------CG--TGTATGTGAAA-------------------TTGTTATTATTAATAACAAC---------AA---------- |
| droBia1 |
scf7180000301345:234620-234836 - |
|
AA-CAGATGAAACGCAAAGCAA--A------CAACGG---CAACAA---CAAAGGCATG----AACAG---GAGCA------------ACAATAA-TTTTAATAA------------CGTTAA-------TAGCCGCAAT--TA--------------------------------------------------------CAATTACAGT---TACAAT---------------AACCAAC------------------------------------------------------------------------------------------------------AACAGTTA------TCGTTAGAGTTACC------------------------------------------------------------------G---T---TACA--CTTACCATTACGATTA--------------CGAGTCCCGTT-----TTTTTTCCAG-TGCGTG------CG--TGTATGTGCAA-------------------TTGTTATTATTAACAACAAC---------AA---------- |
| droTak1 |
scf7180000415281:386829-387062 + |
|
AA-CAGATGAAACGAAAAACGACAA------C---AACAACAACAA---TAGAGGCATG----AACAG---CAGCA------------ACAATAA-TATTAATAA------------CGATAA-------TAGCCACAAT--TACAAT-----------------------------------------------TACAGTTACAATT---------------------ACAATTACCAAC------------------------------------------------------------------------------------------------------AACAGTTA------TCGTTGAAGTTACCG---------------------------------------------------------------CTG---T---TACA--CTTACAATTACGTTCA--------------CGAGTCCCGTT-----TTTTTTCCAG-TGCGTG------CG--TGTATGTGAAA-------------------TTGTTATTATTAACAACGACAA---CAACAA---------- |
| droEle1 |
scf7180000491272:166809-167016 - |
|
A--CAGATGAAACGAAAAACAACAA---------------TAACAA---CAAAGGCATGAATAAACAGCGTCAGCA------------ACAGTAATTTTTAATAT-----------CCGATAA-------TAGCCACA-----GCA--------------------------------------------------ACAACAATTACA---------AC----------------------------------------------------------------------------------------------------------------------------AAC------------------AGTTACC------------------------------------------------------------------G---T---TAAA--CTTACAATTACGTTTA--------------CGAGTCCCGTT----TTTTTTTCCAG-TGCGTG------CG--TGTATGTGAAT-------------------TTGTTATTATTACCAATAACAGAAGCAA-AA---------- |
| droRho1 |
scf7180000779588:305491-305698 + |
|
AA-CAGATGAAACGAAAAACTACAA------CAA------CAACAA---AAAAGGCATGCATGAACAGCGTCAGAA------------ACAGTAT-TTTTAATAT-----------CCGATAA-------TAGCCACA-----GCA--------------------------------------------------ACAACAATTACAAT-------------------------------------------------------------------------------------------------------------------------------------AACAGTTA------TC------------------------------------------------------------------------------G---T---TACA--CTTACAATTACGTTTA--------------CGAGTCCCGTT-----T-TTTTCCAG-TGCGTG------CG--TGTATGTGAAA-------------------CTGTTATTATTAACAATAACAACAACAAC------------ |
| droFic1 |
scf7180000454045:376810-377027 + |
|
AA-CAGATGAAACGAAAAACAGCAA---------------CAACAA---CAAAAGCATG----AACAG---CTGCA------------AGCA-------CATTAACAAATAAAAAAAACATAA-------TAGCCACAGG--A-----------------------------------------------------ACAACAATTACAA-------------------------------C------------------------------------------------------------------------------------------------------AACTGTTA------TCGTTACAGTTACC------------------------------------------------------------------G---T---TACA--CTTACAATTACGTTTA--------------CGAGTCCCGTT-----TTTTTTACAG-TGCGTG------AG--TGTATGTGAAA-------------------TTGTTATTATTAACAACAACAACGGCAACAA---------- |
| droKik1 |
scf7180000302544:951017-951315 + |
|
GG-CATGAACAGCAGC-AGCAAAAATTATAACAACGGCAGCAACAA---CAGCGA----------------CAGCA------------ACTATAG-TTACAATAA------------CAATAA-------GAAATACGAT--AACAATTACAACAATAACACCTGCGGTCACCGCTATATTCACAGTTACAGTCC-ACG-------TT---------------------ACAGTTCCCTAT------------------------------------------------------------------------------------------------------A-TCGTTATAATTCGCGT----------------------------------TACC--------------------------------GTTACCG---T---TACA--CGTACAATTACGTTTACCGCAATCGAAAAGCGAGCTTCAAT-----TTTTTTTCAG-TGCGTG------CG--TGTCTGTGTGCTCCGCCCCAGTCGGTGAAATATCTATT-----------TAGAAATCCTTT---------- |
| droAna3 |
scaffold_13047:1362607-1362858 + |
|
A--CAAACACAACAAC-AACAACAACAACAACAGCAGCAGCAGCAA---CAGAAACAAG----GACAA---CAACA------------ACAACAA-CTGCAACAA------------CAAGAA-------TGGCAACAACATAACAAA---ACAAAAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAACAACGACAAACGCAACAAAACGCAAATT--AACA--------------AC---A---TACA--CTCACAGTT----TTT--------------CAGATTCAGAT-----ATTTGGAGACGGGCCCAAAGGAGTCCACAGAGAT---G-------------------TTGGTACTACAAACAACAACAACAACAACGAGGACAGCGAT |
| droBip1 |
scf7180000396434:619920-620125 - |
|
AA-CAACAAAAACGA----CAAAAATTACAACAACAACAAACACAA---AAATTACAAA----AACAA---CAACA------------AAAACAA-CAACGACAA------------CAAAAA-------CAACAACAAA--AACAAC-----------------------------------------------AACAACAATAAAAAT---TACAACAACA------------------A----------------------------------------------------------------------------CAACAAAAATTAC---AACAAC---AACA------------------------ACA----AAAATTACAACAACAACAAAC--------------------------------ACAAAAA---T---TA------------------------------------------------------------------------------------------------------------------CAAAAACAACAACAAGAACAACAA---------- |
| dp5 |
XR_group8:5517063-5517299 - |
|
GCACACACGGAAGGCGAGACGACGA------C---AA---ACACACAAACACACTCGTA----CAATC---GAGCG------------ACAGAGA-GACATACAC------------CGACAC-------A----------------------------------------------------CACCGACAGGCG-ACG-------AC---------------------GTGGCCCC-CGT------------------------------------------------------------------------------------------------------T-TCCCGAGAGATCGTGT------GAGC----------------------GTCGCT-------------GTCGACGTCGACG----------TAC---A---TACG--GTTACCGTTTCGTTCC--------------CG-TTCCGATC-----GATCGTTCAA-AAA---------AT--CGTATATT-AT-------------------TTATTATTCTTAACAACAAAAATAATAACA----------- |
| droPer2 |
scaffold_10:3253848-3254071 - |
|
CAACA-------------------GCAACAGCAGCAGCTACAACTA---CAGCTACAGC----TACAG---CAGCT------------ACAAGAA-CAACAAGAA------------CAACAA-------CAACAACAGC--TACAGC-----------------------------------------------TACAGCAA-TACAAT---AACAATTTCT------------------A-------------------------------------------------------------------------------------------------------CAGCCA------AAACTATAAGTAAG--TTTT-AGA------------GT-------------CACTGGCT--GTCCACGGTCTCTCTCTCTC---T---CTCT--CTCTCTCTCTCTCCCA--------------CTCGTTCTGT---------TTCTCTG-TGTCTG------TG--TCTGTGTCTA--------------------------------------------------T---------- |
| droWil2 |
scf2_1100000004921:1791829-1792067 - |
|
AA-CAACTACAACAAC-AACAACAACAACAACAACAATTACAACAA---CAACAACAAA----AAT------TACA------AAAACAACAACAA-TTACAACAA------------CAACAA-------CAACCACAAC--AACAAC-----------------------------------------------AACAATAACAACAAC---AACAACAATA------------------A---------------------------------------------------------------------CAACAA-CAACAACAATTAC---AACAAC---AACAACAA------CA------------ACA----ACAATTACA------ACAACA--------------------------------ACAACAACAACAATTA--------------T------------------------------------------------------------------TACAACA-------------------ACAACAATTACAACAACAACAAAAACAGCAA---------- |
| droVir3 |
scaffold_12726:1985648-1985891 + |
|
AC--AAGAACAATAAC-AACAGAAACAACAACAGCAACAACAGCAA---CAACAA----------------CAACA------------ACAACAA-CAGCAGCAA------------C------------------------ATCAGC-----------------------------------------------TGCAAAAGTAAAG-C---TAAG---------------CTTT--CG-GACTCTGTCGCTGGCTGGCTGCTGTTGCTCTCGGCATTGCCAGCAGCGTTTATAGCTAACCGGGCATTGC-------CAGACGCAAATGC---C------------------------------------------------------------------------------------------------------G---C---TACA------------------------------------------------------GCAG-CGAACG------CG--TCAATAAC-AA-------------------CTGCCACTAGTACAAATAAGAAGAAGAACAA---------- |
| droMoj3 |
scaffold_6291:1327-1575 - |
|
AA-CAACAACAACAGC-AGCAGCAG------------CAACAACAA---CTACAGCAGC----AGCAG---CAGCAACAACTACAGCAGCAACTG-CTAAAGCAA------------CAACAGAAACTGA-----ATAAC--AATAAC---AACAACAACAGCAGCAGCAGC-----------------------AGCAACAACTACAGC---AGCAGCAGCA---GCAACAACTA----------------------------------------------------------------------------------------------C---A---------------------------GCAGCAGCAGCAGCAG----CAACA------ACAACA--------------------------------GCAGAAA---CTGCTA-----AAGCAACAACA--------------------G-----------------------------------------------AAAC-------------------TGAATAACAACAACAACAACAAAAACAACAA---------- |
| droGri2 |
scaffold_15203:6433933-6434218 - |
|
AA----ATAATGCGAACAACAACAACAACAGCAACGGCAACAACAA---TAACAACAAT----AACAA---CAGCA------------ACAACAA-CAACAACAA------------CAATAA-------CAGCAGCAGT--AGCAGC-----------------------------------------------AGCAGCAACAACAAT---AATAACAATAGCAGTAACAATAT---------------------------------------------------------------------------CAGCTATAAGCAA-CAAAAGAGCATCAGC---AGCA------------------------ACA----TAAGCAACAG----------C--------------------------------AATGCCA---A--TCAGA--ATGGCGATCATGACTA--------------CGAG----GAT-----ATTTATTTGG-TGCGCGAAGAGTCC--CGCAAGCAAAA-------------------TCAGCAACATCATCAACAGCAGCAGCATCAT---------- |