
ID:dsi_18216 |
Coordinate:x:17130442-17130591 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
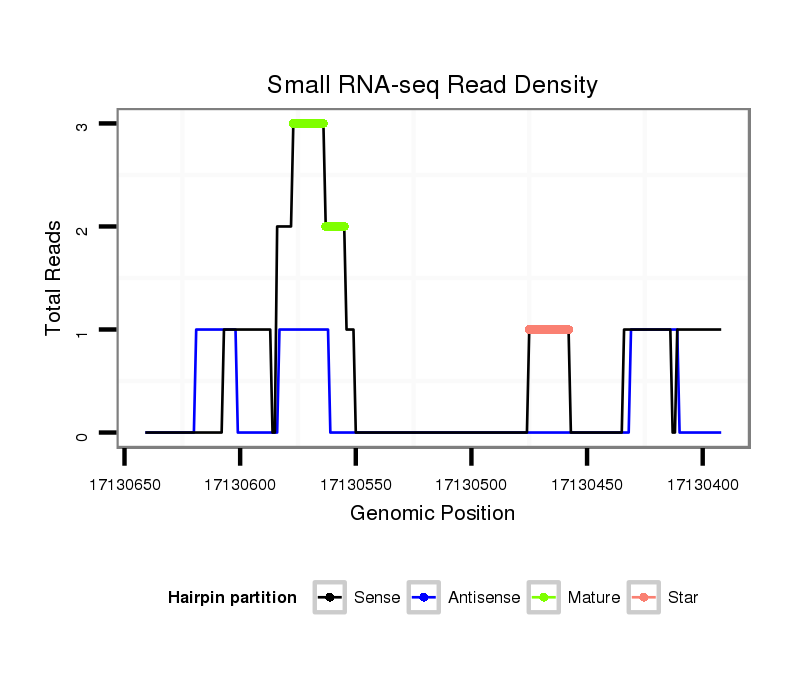
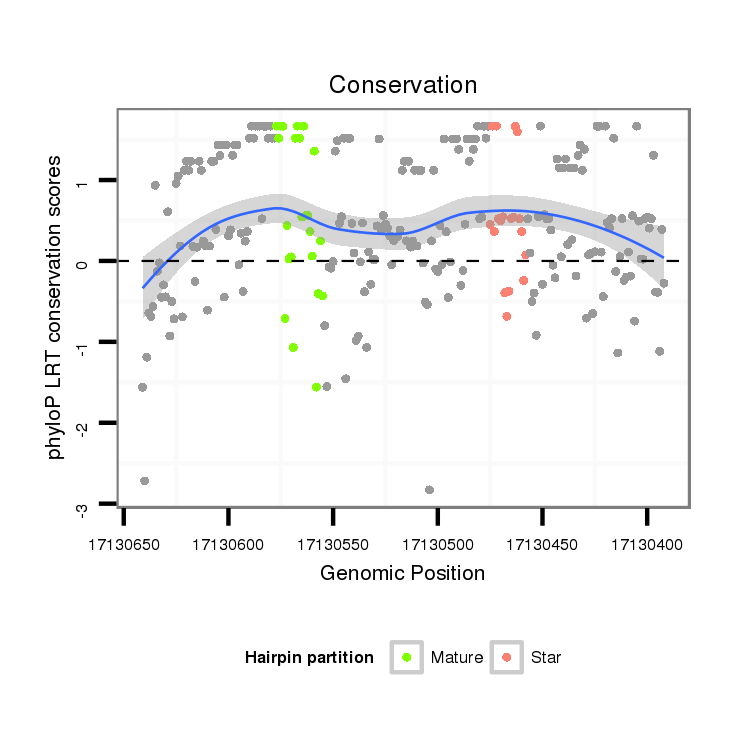
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -24.3 | -24.3 | -24.2 |
 |
 |
 |
five_prime_UTR [x_17130052_17130441_-]; exon [x_17129833_17130441_-]; intron [x_17130442_17130817_-]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TAAACGGTGCTGCGGACACACACATGCACACATTTAACATAGTGTTGTTCGCTGTTAAATCAGCAGTTCGGCCCTGTTAAGCGCACGGGCTAGTGTGGCCGTACTGCTGAAAAATACCGTCCTAGTATGACTGCCAGCAGGGTCTCGTATTGTCATTAGAAAATCTCACATATTCATTTTTCATTTACAAATAAAAATAGCCAGCACACAGCCTACGAATCAGCAGCGCACAAAATAAGTTAGTGTGAAA ****************************************************************.....((((((((..(((......)))....(((((((((((.((........))...)))))))...))))))))))))..............(((((..............)))))..****************************************************************** |
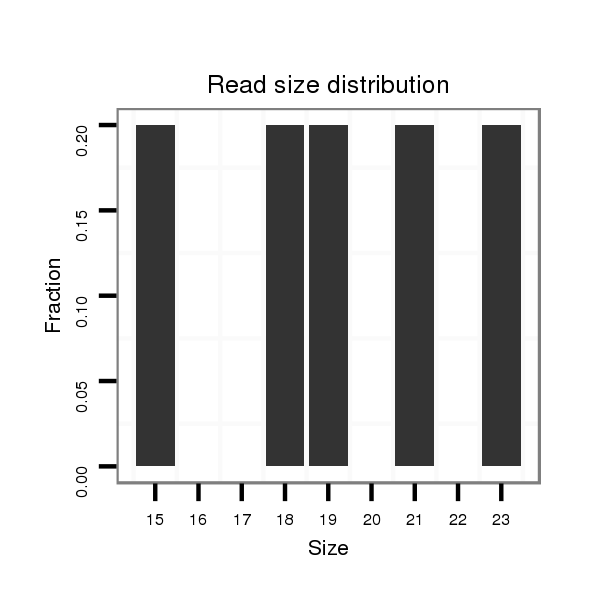
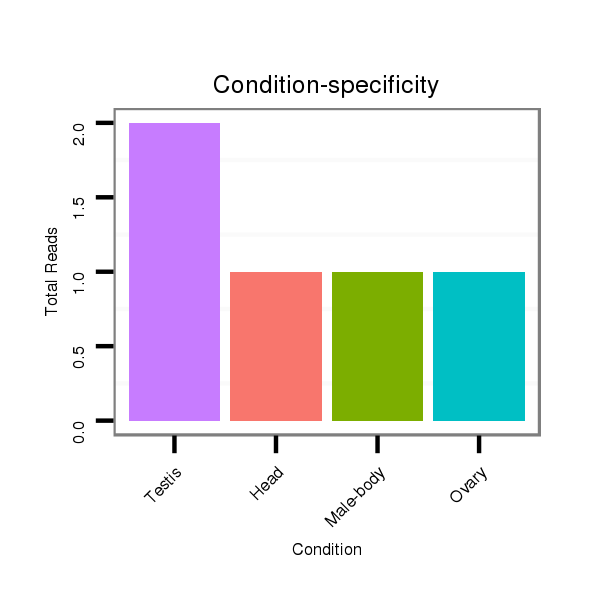
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
M023 head |
O001 Testis |
SRR553485 Chicharo_3 day-old ovaries |
M024 male body |
GSM343915 embryo |
M053 female body |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................ATACCGTCCTAGTCTCA........................................................................................................................ | 17 | 2 | 3 | 6.00 | 18 | 10 | 3 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................TACCGTCCTAGTCTCA........................................................................................................................ | 16 | 2 | 7 | 4.86 | 34 | 16 | 12 | 5 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................ATACCGTCCTAGTCT.......................................................................................................................... | 15 | 1 | 3 | 4.00 | 12 | 8 | 1 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................TACCGTCCTAGTCTCAA....................................................................................................................... | 17 | 3 | 20 | 1.45 | 29 | 8 | 17 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................GATACCGTCCTAGTCTCA........................................................................................................................ | 18 | 3 | 15 | 1.33 | 20 | 8 | 8 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AGTTCGGCCCTGTTAAGCGCACG................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................AATCAGCAGTTCGGCCCTGTT............................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................CAAAATAAGTTAGTGTGAAA | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................ACAGCCTACGAATCAGCAGCG...................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................CCTGTTAAGCGCACGGGCT............................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................GTCCTAGTATCTCTGCCAG................................................................................................................. | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................AATCAGCAGTTCGGC.................................................................................................................................................................................. | 15 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................TAACATAGTGTTGTTCGCTGT................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................CACATATTCATTTTTCAT.................................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................ATACCGTCCTAGTCTCAA....................................................................................................................... | 18 | 3 | 13 | 0.92 | 12 | 9 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................GATACCGTCCTAGTCT.......................................................................................................................... | 16 | 2 | 14 | 0.71 | 10 | 9 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................CGTATTGTAATTAGAACGTCT.................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................AGATACCGTCCTAGTCT.......................................................................................................................... | 17 | 2 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................CTTCTGAACAATACCGTC................................................................................................................................. | 18 | 2 | 5 | 0.40 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ...................................................................................................................ACCGTCCTAGTCTCA........................................................................................................................ | 15 | 2 | 20 | 0.30 | 6 | 2 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................CAGATACCGTCCTAGTCT.......................................................................................................................... | 18 | 3 | 20 | 0.25 | 5 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..AACGGTGCTCCGGAC......................................................................................................................................................................................................................................... | 15 | 1 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..AACGGTGCTCCGGACAG....................................................................................................................................................................................................................................... | 17 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................GATACCGTCCTAGTC........................................................................................................................... | 15 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GTTAAGGGGACGGGTTAGT............................................................................................................................................................ | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................GTATTGTAATTAGAACGTCT.................................................................................... | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................AGATACCGTCCTAGTCTC......................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................TATCGTATTGTAATTAGAAC........................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TCCGCAGTTCGGTCCT............................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................AATATTCAATCTTCATTTACA............................................................. | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................AGATACCGTCCTAGTC........................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
ATTTGCCACGACGCCTGTGTGTGTACGTGTGTAAATTGTATCACAACAAGCGACAATTTAGTCGTCAAGCCGGGACAATTCGCGTGCCCGATCACACCGGCATGACGACTTTTTATGGCAGGATCATACTGACGGTCGTCCCAGAGCATAACAGTAATCTTTTAGAGTGTATAAGTAAAAAGTAAATGTTTATTTTTATCGGTCGTGTGTCGGATGCTTAGTCGTCGCGTGTTTTATTCAATCACACTTT
******************************************************************.....((((((((..(((......)))....(((((((((((.((........))...)))))))...))))))))))))..............(((((..............)))))..**************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
SRR553488 RT_0-2 hours eggs |
M023 head |
SRR553487 NRT_0-2 hours eggs |
M025 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ......................GTACGTGTGTAAATTGTA.................................................................................................................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................CGGATGCTTAGTCGTCGCGTG................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................TAGTCGTCAAGCCGGGACAATT.......................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................GGGGATGCTTAGTCGTCG....................... | 18 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................AAGGTCGTCACAGAGCAT..................................................................................................... | 18 | 2 | 6 | 0.33 | 2 | 0 | 0 | 0 | 2 | 0 |
| ..........................................................................................................................................................................................TGTTTATTTTTATTTGTCGGGTG......................................... | 23 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................................................................................................TGTGAAAGTAAAAAGTAACTGT............................................................. | 22 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................ATCAAAACAAGCGACACT................................................................................................................................................................................................. | 18 | 2 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................TCTATGGCAGGATCAGAG......................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | x:17130392-17130641 - | dsi_18216 | TAA-ACGGTGCTGCGGACA--CACACATGCACA--CATTTAACATAGT-----------GTT-------------GTT-----------CGCTGTTAAATCAGCAGTTCGGCCCTGTTAAGCGCA---------CGG-GCTAGTGTGGCCGTACTGCTGAAAAATACCGTC-CTAGTATGACTGC-------------CAGC-AGG--GTCTCGTATTGTCATTAGA------AAA-TCTCACATATTCAT-------TTTTCA----TTTA----------CAA--ATA--AAAA----TAGC--CAGCACACAGCCTACGAATCA-------GCAGCGCACAAAAT---AAGTTAGTGTG----------------------------------------AAA |
| droSec2 | scaffold_8:340749-340998 - | TAA-ACGGTGCTGCGGACA--CACACATGCACA--CATTTAACATAGT-----------GTT-------------GTT-----------CGCTGTTAAATCAGCAGTTCGGCCCTGTTAAGCGCA---------CGG-GCTAGTGTGGCCGTACTGCTGAAAAATACCGTC-CTAGTATGACTGC-------------CAGC-AGG--GTCTCGTATTGTCGTTAGA------AAA-TCTCACATATTCAT-------TTTTCA----TTTA----------CAA--ATA--AAAA----TAGC--CAGCACACAGCCTACAAATCA-------ACAGCGCACAAAAT---AAGTTAGTGTG----------------------------------------AAA | |
| dm3 | chrX:18031034-18031280 - | TTT-A------------CA--CACACATGCACA--CATTTAACATAGT-----------GTT-------------GTT-----------CGCTGTTAAATCAGCAGTTCGGCCCTGTTAAGCGCA---------CGG-GCCAGTGTGACCGTTCTTCTTAAAAATACCGA-ACCAGTATGACTGCGCTG-C---GACCGTGC-AGG--GTCTCGTATTGTCATTAGA------AAA-TCTCACATATTCAT-------TTTTCA----TTTA----------CAA--ATA--AAAA----TAGC--CAGCACGCAGCCTACAAATTA-------GCAGCGCACAAAAT---AAGTTAGTGTG----------------------------------------AAA | |
| droEre2 | scaffold_4690:8359091-8359356 - | AAA-ACAGTGC---GGGCA--CACACATGCACA--CATTTAACATAGT-----------GTTATTTTGGC----TTTT-----------CGCTGTTAAATCAGCAGTTAGCCCCTGTTAAGCGTG---------CAG-GCCAGTGTGGCCGTCCTGCTGAATAATACCGCCGCCAGTATGACCGCGCCA-C---GGCCCAAA-AGG--CTCTCCTATTGCCATTAGA------AAA-TCTCACATATGCAT-------TTTTCA----TTTA----------CAA--ATT--AAAA----CAGC--CAGCACGCAGCGTTCAAATCA-------GCAGAGAATAAAAC---AAGTTAGTGTA----------------------------------------CAA | |
| droYak3 | X:16654050-16654318 - | TAA-ACGGTGT---GGGCACAC-CACATAGACA--CATTTAACATAGT-----------GTTATTCTGGC----TTTT-----------CGCTGTTAAATCAGCAGTTTGCCCCTGTTAAGCGTG---------CAT-ATCAGTGTGGCCGTCCTGCTGAAAAATACCGCCGCCAGTATGACCGCGACA-C---GACTCAGC-AGG--CTCTCCTATTGTCATTAGA------AAA-TCTGACATATGTAT-------TTTTCA----TTTA----------CCA--ATAAAAAAA----CAGC--CAGCACGCAGCGTTTCAACCA-------GCAGAGAATAAAAC---AAGTTAGTGCA----------------------------------------CAA | |
| droEug1 | scf7180000408176:648285-648558 + | ATA-AGGGCGC-ACATACA--TGCACATGCACA--CCGTTTACAGAGT-----------GTTATTTTGGC----TTTT-----------GGCTGTTAAATCAGCAGTTCTCTTCTGTTAAGTGAA---------CAGGGGCAGTGTGACCGCTTTGTCG-ATAATACCGCCGACAGTATGACCGCGCTAG-CCAGAGCCAAG-AGG--CTCTTTGATTGCTATTAGA------AAA-TCTCACATATGTAT-------TTTTCT----TTTATCCC-------CA--ATA--AAAA----CAGC--CAACACGCAGCGTTTAAATCA-------GCAGCGAAGTAAAC---AAGTTAGTGTT----------------------------------------AAG | |
| droBia1 | scf7180000301760:570540-570809 + | TGCAGGGG-----CGCACA--CGCACACACACG--CTGTTAACAGAGT-----------GTTATTTTGGC----TTTCAAAGGCAATGGCGCTGTTAAATCAGCAGTTCGCCCCTGTTGAGT----------------------GTGACCGCAGCGCG--GAAATGCCGCCGCCAGTGTGGCCGCGCAG-C---TGGCCG-CAGGC--CTGTCCTATTGTCATTAGA------AAA-TCTCACATAGTACTTCTCTATTTTTCC----CTCC-CC-------CAC--ACA--AAAC----CAG---CAGCTCGCAGCGCTAAAACCA-------GCAGCTAATTAGAC---AGGCTAGTGTC----------------------------------------GGG | |
| droTak1 | scf7180000415843:445010-445275 + | CCA-C---------CCACA--CACACACACACA--CGGTTAACAGAAT-----------GTTATTTTGGC----TTTG----------GCGCTGTTAAATCAGCAGTTCGCCCCTGTTAGGT----------------------AGGACCGTTCTGCCG-AAAATACCGCCGCCAGTATGACCGCGCCAG-CT-GAA-CG-A-AGGCCCCATCCTATTGTCATTAGA------AAATTCTCACATATGTAT-------TTTTCT----TTTC-CCCCCTTATTAA--AAA--AAAAAACCCAGCAGCAGCACGCAGCGTTTAAATCA-------GCAGCCAATTAAAC---AGGCTAGTGTT----------------------------------------AAG | |
| droEle1 | scf7180000491193:238717-238974 - | A--------------------CACACATGCACATACAGTTAACAGAGTTCAGGTGCCCTGTTATTTTGGC----TTTT-----------CGCTGTTAAATCAGCAGTTCGCCCCTGTTAAGTGGG---------CAA-CGCAGTGTGGCCATTTTGCC-AAAAATACCGCCGCCAGTATGACCGCGCCAG-CCAGGGCCAAG-AGG--CTCTCCTATTGTCATTAGA------AAA-TCTCACATATGTCT-------TT-TCT----TTTCCATT-------CA--ATA--AAAA-----------------CAGCGTTAAAATAA-------GCAGTTAATTGCTA---AATTTAGTGAT----------------------------------------AAG | |
| droRho1 | scf7180000779986:486078-486340 + | ATG-------C---ATG----CACACATGCACATTCAGTTAACAGAGTTCAGGTGCCCTGTTATTTTGGC----TTTT-----------CGCTGTTAAATCAGCAGTTCGCCACTGTTAAGTGGG---------CAG-GGCAGTGTGGCCGTTTTGCC--AAAATACCGCCGCCAGTATTACCGCGCGAG-CCAGGGTCAAA-AGG--CTCTCCTATTGTCATTAGA------AAA-TCTCACATATGTAT-------TTTTCT----TTCC-CCC-------CA--ATA--AAAA-----------------CAGCATTAAAATCA-------GCAGTGAATTTTAA---AAGTTAGTGTT----------------------------------------AAG | |
| droFic1 | scf7180000453872:320794-321076 + | AAT-AC--------GTGCA--CCCACATGCACACATAGTTAACATAGTGCAGGTGCCCTGTTATTCTTGC----TTTT-----------CGCTGTTAAATCAGCAGTTAGATTCTGTTAAGTGAA---------CAG-AGGGGTGTGACCGTACTGTG-TAAAATACCGCCGCCAGTATGACCGCGCTG--ACAGAGCCC-A-AGG--CTCTCCAATTGTCATTAGA------AAA-TCTCACATATGTAT-------TTTTCT----TTTC-CCC-------CA--A-A--AAAA----CAGCAGCAGCACACAGCATTAAAATCA-------GCAGCAAATTAAAA---AAGTTAGTGAT-------AAGTGT---------------------------GAA | |
| droKik1 | scf7180000302592:1954209-1954457 - | GCA-GGCGCAAC----------------------------AACAAAGCACAAGTGCACTGTTAGTTAGCAGTTCTGGC-----------CCCTGTTAGATCAGCAGTTCGCCTCTGTTAACAGTG---------GGC-AGCAGTGTGCCCGTTTCGG--------ACCACAACCT----------------------------------------------------------ATA-TCTCAGAAATCCGA-------TTTCGA----ACTG----------TAAAAATA--TTTC----AAGG--CAGCACGCA---ATAAAATTA-------GATGCGATTAAAACTAAAAACAAGCGTTTAGAGTTCAGTGCAACTAGTGGAACAAAATTCGCAGTATACAA | |
| droBip1 | scf7180000396362:633299-633484 + | ------------------------------------------------------------------------------------------GCTGTTAAATCAGCAGTTTGCCACTGCTAAGTGTGACCCTGTCGCCC-TGCTGTGTGGCCGTTC-GCTGGGAGAGATA----------------------------GCTA-CA--T--GTCTCATATTGAGAT-AGAAAAAAAAAG-TCTCACATCTGTGT-------ATTTCTTGCCTTTA-CCC-------CA--ACA--AA----------------------CATCAAAATTAGGTAGCAGCAG--CAACAAAT---AAGTTTTT---------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
Generated: 05/18/2015 at 05:39 PM