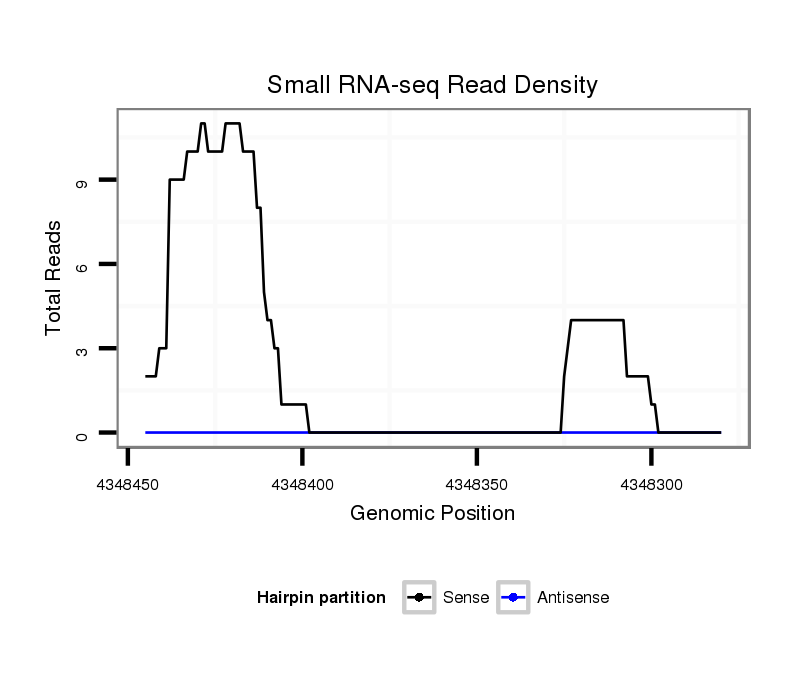
ID:dsi_18192 |
Coordinate:3r:4348330-4348395 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [3r_4348233_4348329_-]; CDS [3r_4348233_4348329_-]; CDS [3r_4348396_4348498_-]; exon [3r_4348396_4348498_-]; intron [3r_4348330_4348395_-]
No Repeatable elements found
| ##################################################------------------------------------------------------------------################################################## AACTTTTTGAGCAACGGCAAGAAGATTGTGGGGGTGGCTCTCAATTACATGTAACTCAGTGCCATTGTAATCTTGGGTCTGCATGGGTAATCCACGAATCCTTTCTTCCGTCGCAGGGACGTTGTGAAGGCCAAGAACGTTCCCGTGCCCAAGGAGCCGCTCGTCT **************************************************..............((((.....((((.......((((.........)))).....))))..))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
M025 embryo |
SRR902009 testis |
SRR618934 dsim w501 ovaries |
M024 male body |
SRR553485 Chicharo_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR902008 ovaries |
M023 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......TGAGCAACGGCAAGAAGATTGTGGGGG.................................................................................................................................... | 27 | 0 | 1 | 3.00 | 3 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .......TGAGCAACGGCAAGAAGATTGTGGG...................................................................................................................................... | 25 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................GTTGTGAAGGCCAAGAAC............................ | 18 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AACTTTTTGAGCAACGGC.................................................................................................................................................... | 18 | 0 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................GATTGTGGGGGTGGCTCTCAATTA....................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TTGTGAAGGCCAAGAACGTTCCCG..................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............AACGGCAAGAAGATTGTGGGGGTGGCT............................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................AAGAAGATTGTGGGGGTGG................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....TTTTGAGCAACGGCAAGAAGATTG.......................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......TGAGCAACGGCAAGAAGATTGTGGGGGT................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................AGAAGATTGTGGGGGTGGTT............................................................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................TGTGAAGGCCAAGAACGTTCCCGTG................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........AGCAGCGTCAAGAAGATTG.......................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................GCAAGAAGATTGTGGGGGTGGCT............................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................GAGGTTGTGAAGGGCAAGA.............................. | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................GCAAGAAGATCGTGGGGGC................................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............GGCAAGAAGATTGTAGGA..................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................CATGTTACTCAGGGCCCT..................................................................................................... | 18 | 3 | 19 | 0.16 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 |
| .........................................................................................................................TTGTGAAGGCCAAAAA............................. | 16 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
|
TTGAAAAACTCGTTGCCGTTCTTCTAACACCCCCACCGAGAGTTAATGTACATTGAGTCACGGTAACATTAGAACCCAGACGTACCCATTAGGTGCTTAGGAAAGAAGGCAGCGTCCCTGCAACACTTCCGGTTCTTGCAAGGGCACGGGTTCCTCGGCGAGCAGA
**************************************************..............((((.....((((.......((((.........)))).....))))..))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
M053 female body |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................AGGCAGCATCCCTGTAACAGT....................................... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........CTCGATGGCGCTCTTCTAACA......................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................GGTTCTGGCAAGGGCACC.................. | 18 | 2 | 6 | 0.33 | 2 | 0 | 0 | 2 | 0 |
| .....................................................................................................................................TCTTGCGAGGGGATGGGTT.............. | 19 | 3 | 14 | 0.14 | 2 | 0 | 2 | 0 | 0 |
| ...............................................................................ACGCACGCATTAGGTGC...................................................................... | 17 | 2 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................TTCTTGGTAGGGAACGGGTT.............. | 20 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 |
| ...............................................................................ACGTACGGATTAGGTGC...................................................................... | 17 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| .................ATTCTGCTAACACCCCCGCCG................................................................................................................................ | 21 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................TCCGGTTGTTGAATGGGCA.................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ............................ACCCCGACCTAGAGTTA......................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| .......................................................................................................AGAAGGCACAGTCCCT............................................... | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
Generated: 04/24/2015 at 02:08 AM