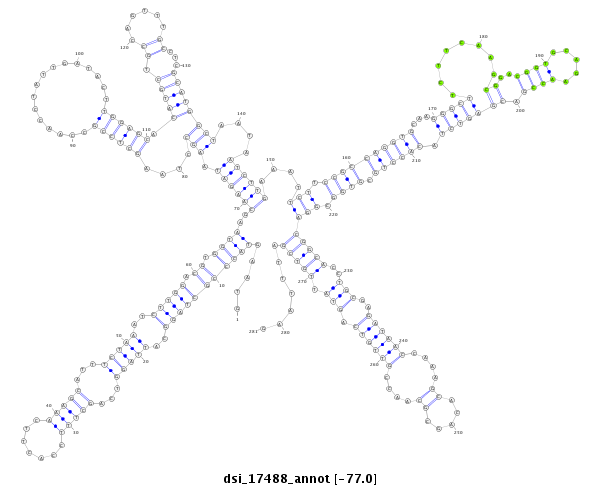
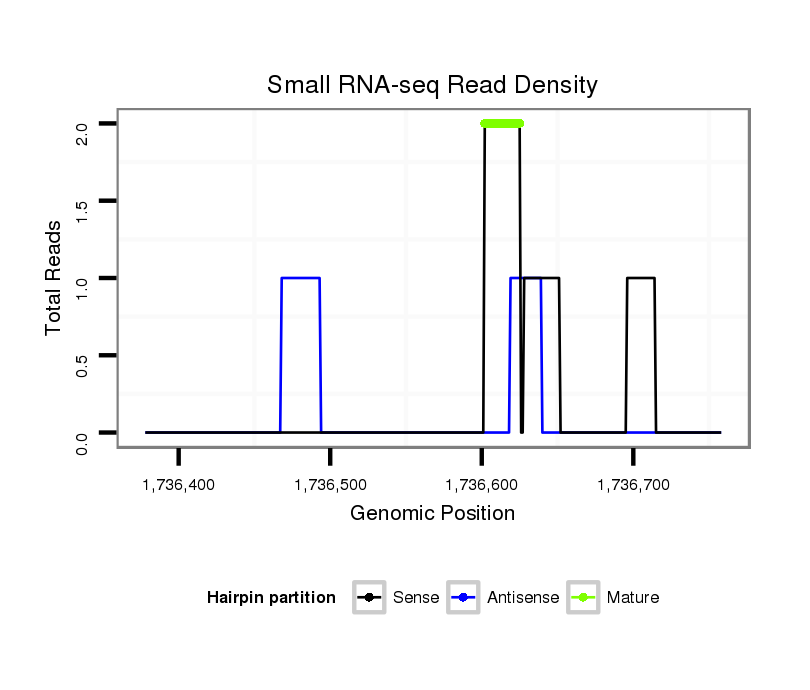
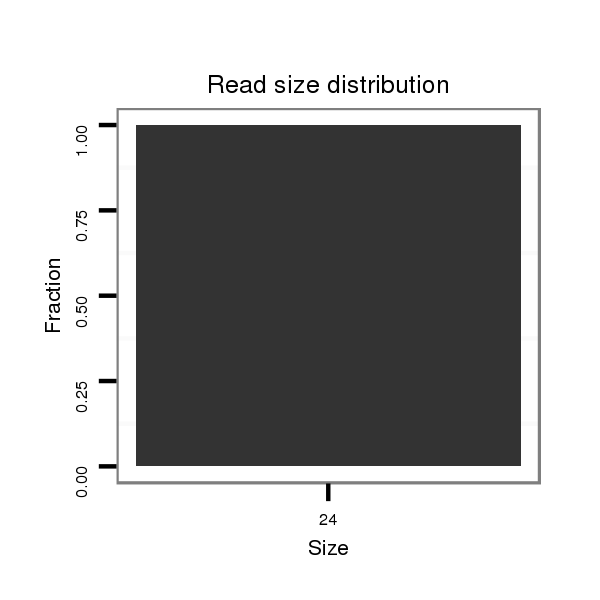
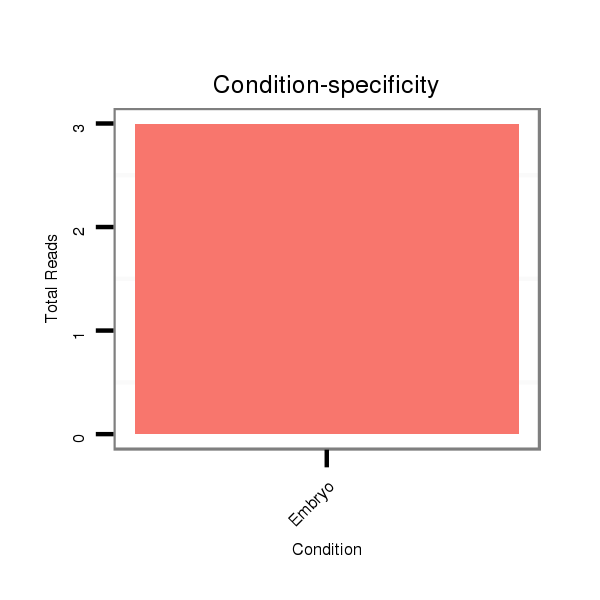
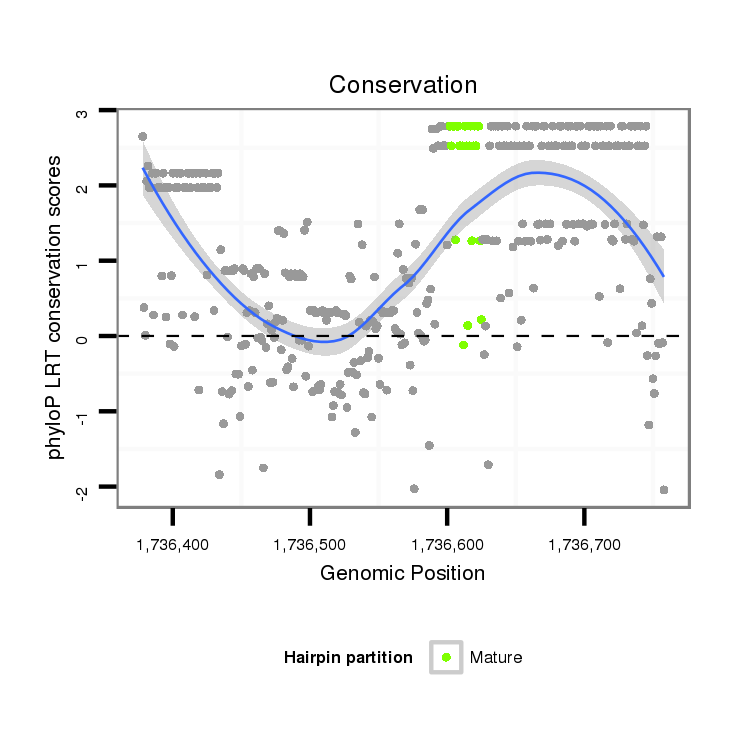
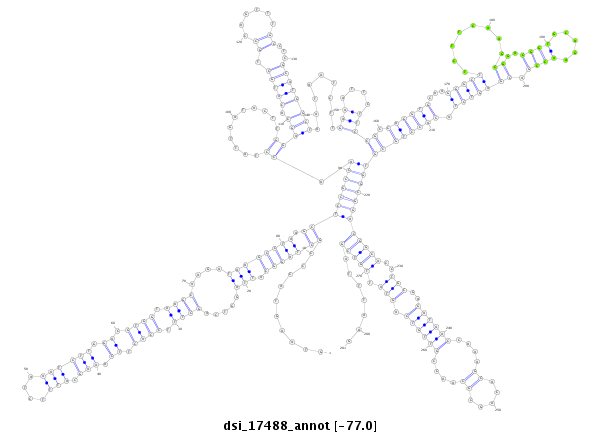
ID:dsi_17488 |
Coordinate:x:1736428-1736708 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -77.0 | -77.0 | -77.0 |
 |
 |
 |
intergenic
No Repeatable elements found
|
TGCGAGGACAAGGCCACCGGCCTGCACTACGGCATCATCACCTGCGAGGGGTAAGTACCCGCTAGGCATTAGGTCAGCTTTCCACTTCAAAGCATTTCTAAATCTTGGACGTGGTAAGCAAGATAAGCCTAAGCTCCGCCAACCTATTGATACTTGGAGCACATGCTGCCAGTTTGCCTCGCATGGCTAATAATCTTGAAATTCTTCCGCCAGGTGCAAGGGCTTCTTCAAGCGGACGGTGCAGAACCGACGAGTCTACACCTGCGTGGCGGACGGCACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAGTGCATCGAGCAGGGCATGGTGCTGCAAGGTGAGTAGTTAGTCCCCTG
**************************************************.....(((((((((((..(((((...(((((.......)))))...)))))..))))).)).))))..((((((.(((....((((((................)))))).(((((.((......))...))))))))....))))))....(((.((((((((((...(((((........((..((((.....)))).))))))).)))))).)))).)))(((((..(((..((((((....((.....))....)))))).))).))))).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
M024 male body |
M023 head |
O002 Head |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................................TCTTCAAGCGGACGGTGCAGAACC..................................................................................................................................... | 24 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................CGAGTCTACACCTGCGTGGCGGAC........................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................AGAATTTCTACATCTTGGAC............................................................................................................................................................................................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................TACGGCATCATCACCTGCGAGT............................................................................................................................................................................................................................................................................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................AGCGCAACCCTTGTCATT................................................................ | 18 | 2 | 3 | 1.00 | 3 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................TAAGAAAGATAAGCATAGGCT...................................................................................................................................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................GAATTTCTACATCTTGGAC............................................................................................................................................................................................................................................................................... | 19 | 2 | 2 | 1.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................TTGTCGATTTAAGAAGTGC............................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................GCGCAACCCTTGTCATTAGT............................................................. | 20 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................GAGCGCAACCCTTGTCATT................................................................ | 19 | 3 | 12 | 0.25 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................GCGCAACCCTTGTCATTA............................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................CCTGCGAGGGGTAGCTA.................................................................................................................................................................................................................................................................................................................................... | 17 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................AGATTACCAAAGCAC.................................................................................. | 15 | 1 | 13 | 0.15 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ............................................................................................AATTTCTACATCTTGGAC............................................................................................................................................................................................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAGGTCCCCGCCAGGCAT........................................................................................................................................................................................................................................................................................................................ | 19 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................GCGCAACCCTTGTCATT................................................................ | 17 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................GAGCGCAACCCTTGTCA.................................................................. | 17 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................ACCGTTGCTAGTATTGT........................................................... | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................CGCAACCCTTGTCATTA............................................................... | 17 | 2 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................GGTTGTCAGTATTGAGGAT....................................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................AATCCTGGATGTTGTAAGC...................................................................................................................................................................................................................................................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................AAGTGCCCGCTAGGCT......................................................................................................................................................................................................................................................................................................................... | 16 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................CGCAACCCTTGTCATTAGT............................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................CCTGCGAGGGGTAGCT..................................................................................................................................................................................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
ACGCTCCTGTTCCGGTGGCCGGACGTGATGCCGTAGTAGTGGACGCTCCCCATTCATGGGCGATCCGTAATCCAGTCGAAAGGTGAAGTTTCGTAAAGATTTAGAACCTGCACCATTCGTTCTATTCGGATTCGAGGCGGTTGGATAACTATGAACCTCGTGTACGACGGTCAAACGGAGCGTACCGATTATTAGAACTTTAAGAAGGCGGTCCACGTTCCCGAAGAAGTTCGCCTGCCACGTCTTGGCTGCTCAGATGTGGACGCACCGCCTGCCGTGGACGCTCTATTGGTTTCGTGTCGCGTTGGCAACAGTCATAACAGCTAAATTCTTCACGTAGCTCGTCCCGTACCACGACGTTCCACTCATCAATCAGGGGAC
**************************************************.....(((((((((((..(((((...(((((.......)))))...)))))..))))).)).))))..((((((.(((....((((((................)))))).(((((.((......))...))))))))....))))))....(((.((((((((((...(((((........((..((((.....)))).))))))).)))))).)))).)))(((((..(((..((((((....((.....))....)))))).))).))))).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O002 Head |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR1275485 Male prepupae |
SRR553488 RT_0-2 hours eggs |
M023 head |
M025 embryo |
O001 Testis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................................................................GACGATCTATTGGTTTCG.................................................................................... | 18 | 1 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................TTAATTCTTCACGTAGCTCAACC.................................. | 23 | 3 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................GTCTTGGCTGCTCAGATGTGG....................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................TCGTAAAGATTTAGAACCTGCACCAT......................................................................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................CTCTATTTGTTTCGGGTCGCG............................................................................. | 21 | 2 | 2 | 1.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................AGGTGATGCAGTAGTAGTGT................................................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 10 | 0.70 | 7 | 0 | 1 | 1 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................CTCTATTTGTTTCGTG.................................................................................. | 16 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................CTCTATTGGTTTCGGGTCACG............................................................................. | 21 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........TCCTGTGGCGGGACGTGA................................................................................................................................................................................................................................................................................................................................................................. | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................CGATCTATTGGTTTCG.................................................................................... | 16 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................AAGTTTCGTAAAGAGTT....................................................................................................................................................................................................................................................................................... | 17 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................CCGGATGTGATGCCTTTG......................................................................................................................................................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................CCAGTCGAAGTGTGCAGT.................................................................................................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................ACTCTCTATTTGTTTCGGGTC................................................................................ | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................ACCACGAGGTTCCAC................ | 15 | 1 | 18 | 0.11 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................ACAGTCGTAAGAGATAAATT................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................................................................GCACGTCTTGGGTGCT................................................................................................................................ | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................TGCGCCTGCATCGTCTTGG..................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................AGTGATAACAGTGAAATTC.................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................CGTACAACGGCGTTCCGCT............... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......AGGTACCGGTGGCCGGACG.................................................................................................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | x:1736378-1736758 + | dsi_17488 | TGCGAGGACAAGGCCACCGGCCTGCACTACGGCATCATCACCTGCGAGGGGTAAGTA----------------------------------------CCCGCTAGGCATTAGGTCAGCTTTCCACTTCAAAG----CAT-----------------------------TTCTAAAT------CTTGGACGTGGTAA--GCA-AGATAAGCCTAAGCTCCGCCAACCTATTGATACTTG-----------------------GAGCACATGCTGCCAGTTTGCCTCGCATGGCTAAT------AATCTTGA-A--------------------------------------------------------------ATTCTT------------------------------------------------CCG-CCAGGTGCAAGGGCTTCTTCAAGCGGACGGTGCAGAACCGACGAGTCTACACCTGCGTGGCGGACGGCACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAGTGCATCGAGCAGGGCATGGTGCTGCAAGGTGAGTA-----------------GTTA-------------G--TCCCCT---------G |
| droSec2 | scaffold_10:1679978-1680358 + | TGCGAGGACAAGGCCACCGGCCTGCACTACGGCATCATCACCTGCGAGGGGTAAGTA----------------------------------------CCCGCTAGGCATTAGGTCAGCTTTCCACTTCTAAG----CAT-----------------------------TTCTAAAT------CTTGGACGTGGTAA--GCA-AGATAAGCCTAAGCTCCGCCAACCTATTGATACTTG-----------------------GAGCACATGCTGCCAGTTTGCCTCGCATGGCTAAT------AATCTTGA-A--------------------------------------------------------------ATTCTT------------------------------------------------CCG-CCAGGTGCAAGGGCTTCTTCAAGCGGACGGTGCAGAACCGACGAGTCTACACCTGCGTGGCGGACGGCACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAGTGCATCGAGCAGGGCATGGTGCTGCAAGGTGAGTA-----------------CTTA-------------G--TCCCCT---------G | |
| dm3 | chrX:1894389-1894754 + | TGCGAGGACAAGGCCACCGGCCTGCACTACGGCATCATCACCTGCGAGGGGTAAGTA----------------------------------------CGTGCTAGGCATTAAGTCAGCTTTCCACTTCTAAG----CGT-----------------------------TTCTAAAT------CTTGGACATGGTAA--GCA-AGATAAGCCAAAGCTCCGTCAACATC--------------------------------------AATGCTACCACTTTGCCCCGCATGGCTAAT------AATCTTGG-A--------------------------------------------------------------ATTCTC------------------------------------------------CCG-ACAGGTGCAAGGGCTTCTTCAAGCGGACGGTGCAGAACCGACGAGTCTACACCTGCGTGGCGGACGGCACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAGTGCATCGAGCAGGGCATGGTGCTGCAAGGTGAGTA-----------------CTTA-------------G--TCCCCT---------G | |
| droEre2 | scaffold_4644:1864905-1865280 + | TGCGAGGACAAGGCCACCGGCCTGCACTACGGCATCATCACCTGCGAGGGGTAAGTA----------------------------------------CCCGCTGGCCACCGAGTCAGTTTTCCACTTCCAAG----CGT---------------------------------------------TGGACATGGTAAGAGCC-AGATCAGCTTGAGC-CCGTGCACCCATTGCCACCCA-----------------------CAG----------CAGTCTGCCTCGCATGGCTAAT------GATCTTGC-A--------------------------------------------------------------ATTCTC---------------------------CAATTCTCCAAT-TC------TCG-CCAGGTGCAAGGGCTTCTTCAAGCGGACGGTGCAGAACCGACGAGTCTACACCTGCGTGGCGGACGGCACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAGTGCATCGAGCAGGGCATGGTGCTGCAAGGTGAGTA-----------------CTTG-------------G--TCCACT---------G | |
| droYak3 | 3L:15308215-15308368 + | T-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAAGGGCTTCTTCAAGCGCACCGTGCAGAACAAGAAGGTCTACACCTGCGTGGCGGAGCGGTCGTGCCACATCGACAAGACGCAGCGCAAGCGGTGTCCCTACTGCCGATTCCAGAAGTGCCTTGAAGTGGGCATGAAGCTAGAGGGTGAGT------------------------------------------------------ | |
| droEug1 | scf7180000409182:437739-438037 + | TGCGAGGACAAGGCCACCGGCCTGCACTACGGCATCATCACGTGCGAGGGGTAAGTAAAAAGCTTCCTGAGTC-------------------------------------------------------------------------------------------------CTAAAT------C----------------CA-AA--------------------------------CT-----------------------GAGTACTAA----------------------T-CTAATCCGAATCCTTGAA--------------------------------------------------------------ATTTT-------------AC-----------------------------CT---CCG-TCAGGTGCAAGGGCTTCTTTAAGCGGACGGTGCAGAACCGACGCGTCTACACCTGCGTGGCGGACGGCACCTGCGAGATAACCAAAGCACAGCGGAACCGTTGTCAGTATTGTCGATTTAAGAAGTGCATCGAGCAGGGCATGGTGCTGCAAGGTGAGTA-----------------CTTGAC-----------G--TCTCCT---------- | |
| droBia1 | scf7180000301760:4499045-4499225 + | TC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CG-CCAGGTGCAAGGGCTTCTTCAAGCGAACGGTGCAGAACCGACGCGTCTACACCTGCGTGGCGGACGGCACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAGTGCATCGAGCAGGGCATGGTGCTGCAGGGTGAGTACC----GCCGACAGCCGCTT--------------------------------G | |
| droTak1 | scf7180000415211:47286-47456 - | T-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCA-CCAGGTGCAAGGGCTTCTTCAAGCGGACGGTGCAGAACCGACGCGTCTACACCTGCGTGGCGGACGGCACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAGTGCATCGAGCAGGGTATGGTGCTGCAAGGTGAGTG-----------------ATTA-------------T--T--------------A | |
| droEle1 | scf7180000491001:1405952-1406140 + | T-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AATCCTAG-A--------------------------------------------------------------ACTCTC------------TC--------CGAAT---------------GCT---CCG-TTAGGTGCAAGGGCTTCTTCAAGCGGACGGTGCAGAACCGACGGGTCTACACCTGCGTGGCGGACGGCACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAGTGCATCGAGCAGGGCATGGTGCTGCAAGGTGGGTA----------------------------------------------------- | |
| droRho1 | scf7180000777610:24110-24300 - | TGAGA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TC--------------------------------C-----------ATCTA-ATC---CCCAGGTGCAAGGGCTTCTTCAAGCGGACGGTGCAGAACCGACGGGTCTACACCTGCGTGGCGGACGGCACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAGTGCATCGAGCAGGGCATGGTGCTGCAAGGTGAGTG------GG---------CGGA-------------A--TCTCCT---------C | |
| droFic1 | scf7180000454072:3007055-3007236 + | TC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CG-CCAGATGCAAGGGCTTCTTCAAGCGGACGGTGCAGAACCGACGCGTCTACACCTGCGTGGCGGACGGCACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAGTGCATCGAGCAGGGCATGGTGCTGCAAGGTGAGTG-----------------TT------------------CCTCCTCCTCCTCCTA | |
| droKik1 | scf7180000302610:159733-160030 + | TGTGAGGATAAGGCCACCGGCCTTCACTATGGCATCATCACGTGCGAAGGGTGAGTGTTGTTATTTTGGATTC-------------------------------------------------------------------------------------------------AAAAAT------C----------------CC-AA--------------------------------TT------------------CCCTGGAGTATGTA----------------------T-ATATTT------T---------------------------------------------------------------CTATATTTTT------------------------------------------------CTGATCAGGTGCAAGGGCTTCTTCAAGCGGACGGTGCAGAACAGGCGTGTCTACACCTGCGTGGCGGATGGAACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAGTGCATCGAGCAGGGCATGGTGCTGCAAGGTGAGTG-----------------GTTAAG-----------GTTGCTCCC---------T | |
| droAna3 | scaffold_12929:1886070-1886348 + | TGCGAGGACAAGGCCACCGGTCTCCACTACGGCATCATAACATGCGAGGGGTAAGTCTTCCCA----------C----------------------------------------------------------------------------------------------------C-TTCTTGC----------------CACAA--------------------------------CTGGGTAGAGTATACTAATGTCCT-----------------------------------------G-------------------------GATCCTT-------------------------------------------------------------------------------------------------TGCAGGTGCAAGGGCTTCTTCAAGCGGACGGTGCAGAACCGGAGGGTCTACACCTGCGTGGCGGATGGCACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGTTTTAAGAAGTGCATCGAGCAGGGCATGGTGCTGCAGGGTCAGTA-----------------TT------------------CCTTCT---------A | |
| droBip1 | scf7180000396543:346817-347103 + | TGCGAGGACAAGGCCACAGGCCTCCACTACGGCATCATAACATGCGAGGGGTAAGTTCCA-----------------------------------------------------------------------C----CACTAGCTTCTCCAGTACTCTGGATCGTAATCG------------------------------------------------------------------------------------------------------------------------------------G-------------------------GATCTAT-------------------------------------------------------------------------------------------------TGCAGGTGCAAGGGCTTCTTCAAGCGGACGGTGCAGAACCGAAGGGTCTACACCTGCGTGGCGGATGGCACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGTTTTAAGAAGTGCATTGAGCAGGGTATGGTGCTGCAAGGTCAGTA-----------------TT-GTTGTATTCCATTAT--CCCCCA---------T | |
| dp5 | XL_group1a:2564263-2564567 + | TGCGAGGACAAGGCCACTGGCCTGCACTACGGCATCATCACCTGCGAGGGGTAAGTGTT------------------------------------------------------------TTC-----CCACG----CCT---------------------------------------------ATGAGATGGCAGCAG---------------------------------CACCCA-----------------------CAC----------CAGA-----------GACTA-----------------------------------------------ATCC--TCCCCCCATCGCCCCCACCCTCCCCATA--------------------------------C-----------TTTTC-ATC---CACAGCTGCAAGGGCTTCTTCAAGCGCACGGTGCAGAACCGACGCGTCTACACCTGCGTGGCGGACGGCACCTGCGAAATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAATGCATTGAGCAGGGCATGGTGCTGCAGGGTGAGTA----------------------------------------------------- | |
| droPer2 | scaffold_28:1001201-1001505 + | TGCGAGGACAAGGCAACTGGCCTGCACTACGGCATCATCACCTGCGAGGGGTAAGTTTT------------------------------------------------------------TGC-----CCACG----CCT---------------------------------------------TGGAGATGGCAGCAG---------------------------------CACCCA-----------------------CAC----------CAGA-----------GACTA-----------------------------------------------ATCC--TCCCCCCATCGCCCCCACCCTCCCCATA--------------------------------C-----------TTTTC-ATC---CACAGCTGCAAGGGCTTCTTCAAGCGCACGGTGCAGAACCGACGCGTCTACACCTGCGTGGCGGACGGCACCTGCGAAATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAATGCATTGAGCAGGGCATGGTGCTGCAGGGTGAGTA----------------------------------------------------- | |
| droWil2 | scf2_1100000004909:7007339-7007654 - | TGTGAGGATAAGGCCACCGGATTACACTATGGCATCATTACATGCGAGGGGTGAGTAATA--------------------------CACACAGAGAACTGGCCTGGCATTAGTTCC-------ATTCGT---TTGCAAC----------------------------------------------------------------------------------------------------------------------------------------TGC-----------AACTC-----------------------------------------------TTTTTTTCCTTCTCTCTCTCTCTCTC---------TCTC------------------------------------------------TCG-TTAGCTGCAAGGGATTCTTCAAGCGCACGGTACAGAATCGTCGGGTTTATACCTGCGTGGCGGATGGCACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAATGCATTGAGCAGGGCATGGTGCTGCAAGGTAAGTCCAGAACAACGAA----------------------------------------- | |
| droVir3 | scaffold_12932:1651335-1651617 + | TGCGAGGACAAGGCCACCGGCCTGCACTACGGCATCATCACCTGCGAGGGGTAAGTGTCGCCA----------CAAATCTCCAATTTAAGCAAAAAACTA-------------------------------------AC-----------------------------TGCTAAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCCTC-ACC---TGCAGCTGCAAGGGCTTCTTCAAGCGCACGGTGCAGAACCGCAGGGTCTACACCTGCGTCGCGGACGGCACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAGTGCATCGAGCAGGGCATGGTGCTGCAAGGTCAGTA------GACGAAGCCAACTT--------------------------------G | |
| droMoj3 | scaffold_6473:2957941-2958282 - | TGCGAGGACAAGGCCACCGGACTGCACTACGGCATCATCACCTGCGAGGGGTAAGTTGAA-----------------------------------------------------------------------G----CAC-----------------------------GGCTAAAC------CT---------------CA-AG--------------------------------TG-----------------------GAGCACTTGA--------------------------------------CAAAGTTGATCTAACATTGATCTATCTCCCTCTCTCTCTCTCTCTCTCTCTCTCTCTC---------TCTCTCTCTCCATCTC-CCTCTGTGA-----------------TCCTC-TCC---TGCAGATGCAAGGGCTTCTTCAAGCGCACCGTGCAGAACAGAAGGGTCTACACGTGCGTTGCCGATGGCACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAATGCATTGAGCAGGGCATGGTGCTGCAAGGTCAGTA----------------------------------------------------- | |
| droGri2 | scaffold_15203:5573086-5573244 - | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGCTGCAAGGGCTTCTTCAAGCGGACCGTGCAGAACCGACGCGTCTACACGTGCGTGGCGGATGGAACCTGCGAGATAACCAAAGCACAGCGCAACCGTTGTCAGTATTGTCGATTTAAGAAATGCATTGAGCAGGGCATGGTGCTGCAAGGTCAGTA----------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 10:08 AM