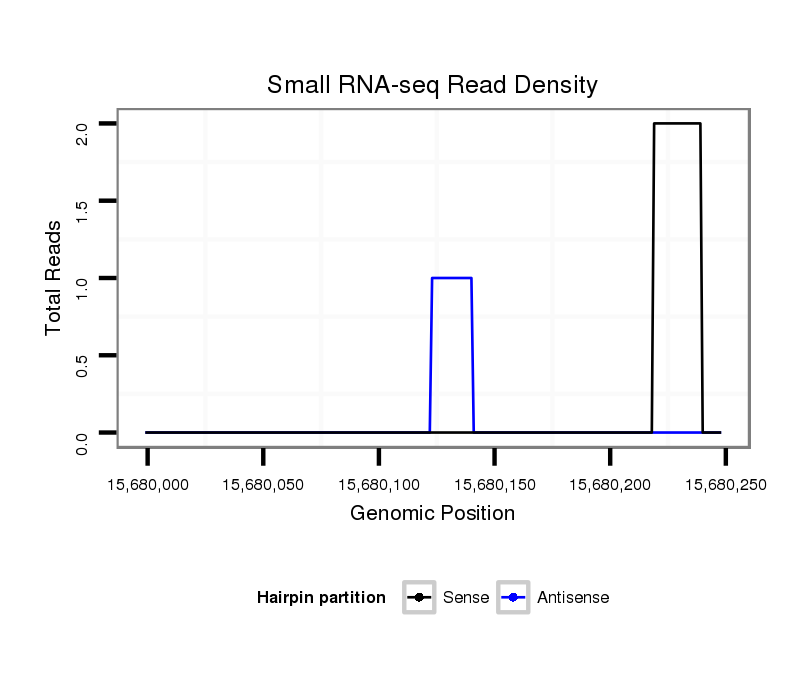
ID:dsi_17346 |
Coordinate:3l:15680049-15680198 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
CDS [3l_15680199_15681029_+]; exon [3l_15680199_15681029_+]; intron [3l_15678936_15680198_+]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGATGATTATCATTTTGCATGTCCCATTTGCATGCGTAATCAAATCGGGTCACCCTAAGTATCATTATAGTATGCAATTGATATTTACATATATGTTATATAAATAGTGAGAAATTATTTGATTTTTGTTTGAAACACACTCGCGTCCATCGCCTTAGGACAACTGTTTTTTATTTGTTCCGAACTAATATTATTAACAGAACGAAACAAGCTCTGCATCATCGACATCTGCTCCAACACCATCGATCCT **************************************************...((((((..(((((((..((((.(((..((((....))))..))).)))).)))))))((((((....))))))((((...)))).....(((.....)))))))))....(((((...((((.(((...))).)))).....)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
SRR902009 testis |
SRR553486 Makindu_3 day-old ovaries |
M023 head |
|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................................ATCGACATCTGCTCCAACACC......... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| .....................................................................................................................................AACCAACTCGCGTCCATCGGC................................................................................................ | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................TATTATTAACAGATCGAAAC.......................................... | 20 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .................CAAGTCCCATCGGCATGCGTA.................................................................................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................ACCAACTCGCGTCCATCGGC................................................................................................ | 20 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................TTCGGACAACTGTATTTTATTTA......................................................................... | 23 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 |
| .................................................................................................................................TTAAAACGCACTCGCGACC...................................................................................................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 |
|
ACTACTAATAGTAAAACGTACAGGGTAAACGTACGCATTAGTTTAGCCCAGTGGGATTCATAGTAATATCATACGTTAACTATAAATGTATATACAATATATTTATCACTCTTTAATAAACTAAAAACAAACTTTGTGTGAGCGCAGGTAGCGGAATCCTGTTGACAAAAAATAAACAAGGCTTGATTATAATAATTGTCTTGCTTTGTTCGAGACGTAGTAGCTGTAGACGAGGTTGTGGTAGCTAGGA
**************************************************...((((((..(((((((..((((.(((..((((....))))..))).)))).)))))))((((((....))))))((((...)))).....(((.....)))))))))....(((((...((((.(((...))).)))).....)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
M024 male body |
SRR553488 RT_0-2 hours eggs |
O001 Testis |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
M023 head |
M053 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................GGAATCCTGTAGACGCAA................................................................................ | 18 | 3 | 20 | 1.75 | 35 | 2 | 27 | 0 | 0 | 2 | 0 | 3 | 1 |
| ............................................................................................................................AAACAAACTTTGTGTGAG............................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................AGCGGAATCCTGTGGACGCAA................................................................................ | 21 | 3 | 3 | 0.67 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TCAGGGGGATTCATAGT.......................................................................................................................................................................................... | 17 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................CGGAATCCTGTGGAC.................................................................................... | 15 | 1 | 5 | 0.40 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................AGCGGAATCCTGTGGACGCA................................................................................. | 20 | 3 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................GGGGAATCCTGTGGAC.................................................................................... | 16 | 2 | 14 | 0.36 | 5 | 1 | 0 | 1 | 0 | 0 | 3 | 0 | 0 |
| ...................................................................................................................................................................................................CTGTCTTGCTGTGTTAGAGACGT................................ | 23 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................AGCGGAATCCTGTGGACG................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................GCGGAATCCTGTGGACGCAA................................................................................ | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................AAAAAATAAAGACGGCTCGATT.............................................................. | 22 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................GAACGTACGCATTAGGT............................................................................................................................................................................................................... | 17 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................GGTTGAACGTACGGATTAG................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
Generated: 04/23/2015 at 05:24 PM