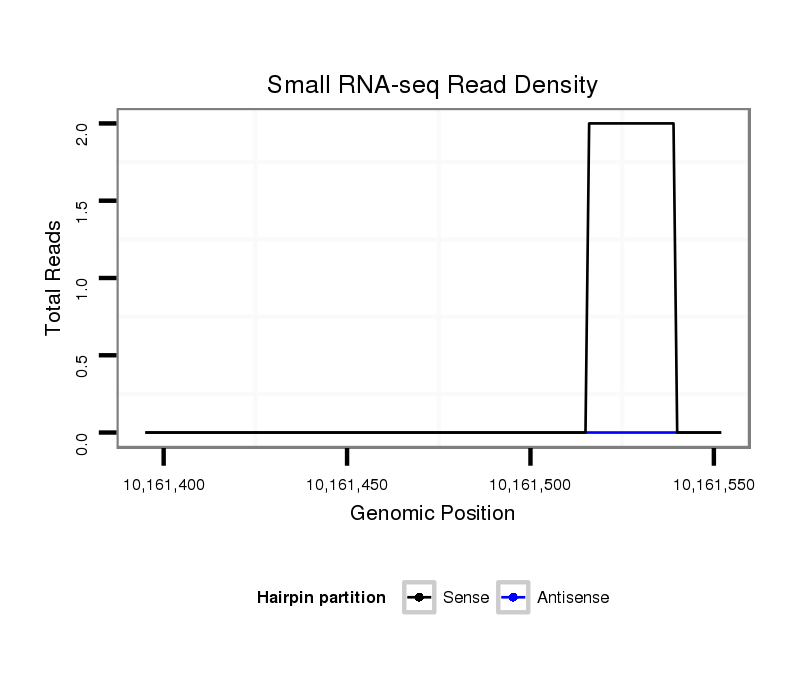
ID:dsi_17311 |
Coordinate:3r:10161445-10161502 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
CDS [3r_10161244_10161444_+]; exon [3r_10161225_10161444_+]; CDS [3r_10161503_10161611_+]; exon [3r_10161503_10161611_+]; intron [3r_10161445_10161502_+]
No Repeatable elements found
| ##################################################----------------------------------------------------------################################################## ATATATCCTGGACCACAATCTTTGAAGCGTCGAAGGAGGCTCTCTTTAAGGTTCCTAGCCATTATCGTTAAACAATCAGTTCCATTTTAATTTTTGCCCTAATAACAGCAAGCTGGAAATCTGGAGGATGTCAACGAGAAAGTATTCAAGACTCTGGC **************************************************..........................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR902008 ovaries |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
M023 head |
M024 male body |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........TGGACCAAAAGCTTTGAAGCGTT............................................................................................................................... | 23 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TGGAGGATGTCAACGAGAAAGTAT............. | 24 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................TAAGCAAGCTGGAAATCTGGAGGATGTC.......................... | 28 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..ATATGTTGGACCACAATCTTTT...................................................................................................................................... | 22 | 3 | 3 | 0.67 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| .......TTGGACCACAATCTTTAAA.................................................................................................................................... | 19 | 2 | 5 | 0.60 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........TGGACCACAATCTTT....................................................................................................................................... | 15 | 0 | 4 | 0.50 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .GATATGTTGGACCACAATCTTT....................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................CGTCGAAGTAGGCTGTCT................................................................................................................. | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TGGAGGATGTCAACGA..................... | 16 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................GTGCCATTTTAGTTTTTGAC............................................................ | 20 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| ........................................................................................................ACAGCAGGCTGGAAAAATGGA................................. | 21 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................GGAGGAGGCTCTCTT................................................................................................................ | 15 | 1 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................AGGATATCAACGAGA................... | 15 | 1 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................TGAAAATCTGGAGGA.............................. | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........TGGACCACAATCTGTC...................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................CTAACCACTATCGTTA........................................................................................ | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................AGTGTCGAAGGCGGCTGTC.................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
TATATAGGACCTGGTGTTAGAAACTTCGCAGCTTCCTCCGAGAGAAATTCCAAGGATCGGTAATAGCAATTTGTTAGTCAAGGTAAAATTAAAAACGGGATTATTGTCGTTCGACCTTTAGACCTCCTACAGTTGCTCTTTCATAAGTTCTGAGACCG
**************************************************..........................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................CAGCTGCCTTCGAGAGAAA............................................................................................................... | 19 | 2 | 2 | 2.00 | 4 | 3 | 0 | 1 | 0 | 0 | 0 | 0 |
| AATATAGGACCTGGTGTTAGAA........................................................................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................ACAGCTGCCTGCGAGAGAAA............................................................................................................... | 20 | 3 | 8 | 0.25 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................TCTTTCATAAGTTGCGAG.... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................GGATTGGTGATAGCAATAT...................................................................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................ACAGCTGCCTTCGAGAGAAA............................................................................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................TCGACCTCTAGACCTC................................ | 16 | 1 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................ACAGCTGCCTTCGAGAGAA................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
Generated: 04/24/2015 at 10:33 AM