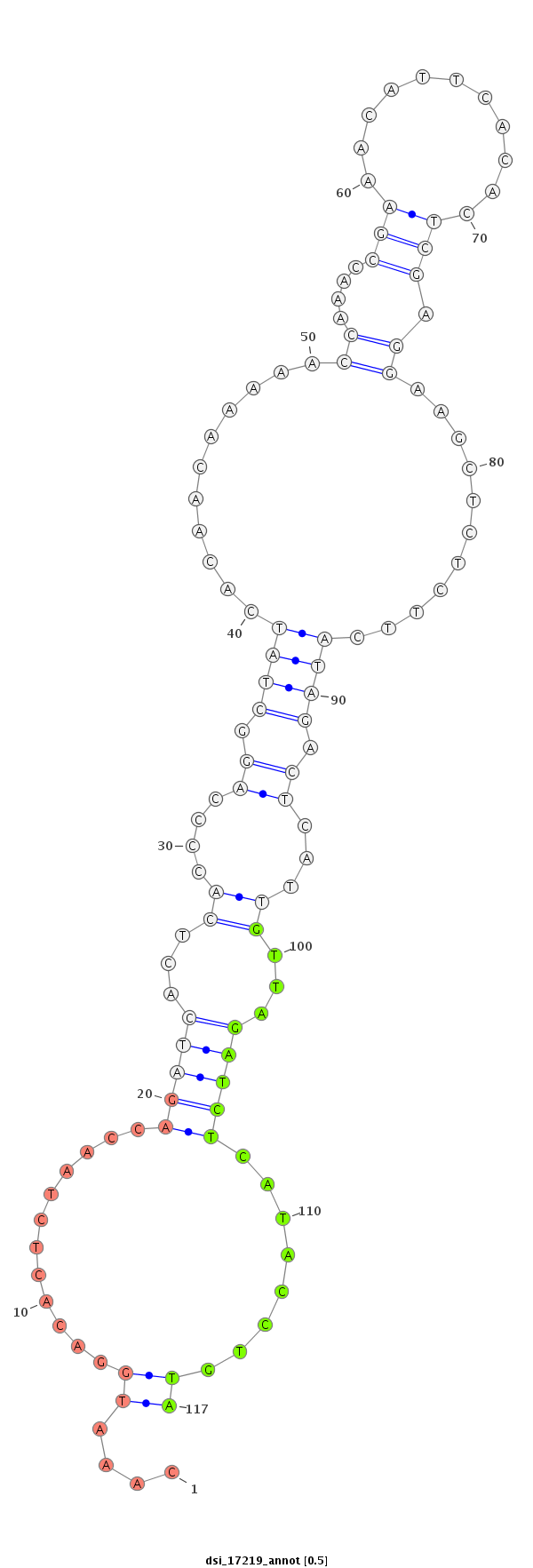


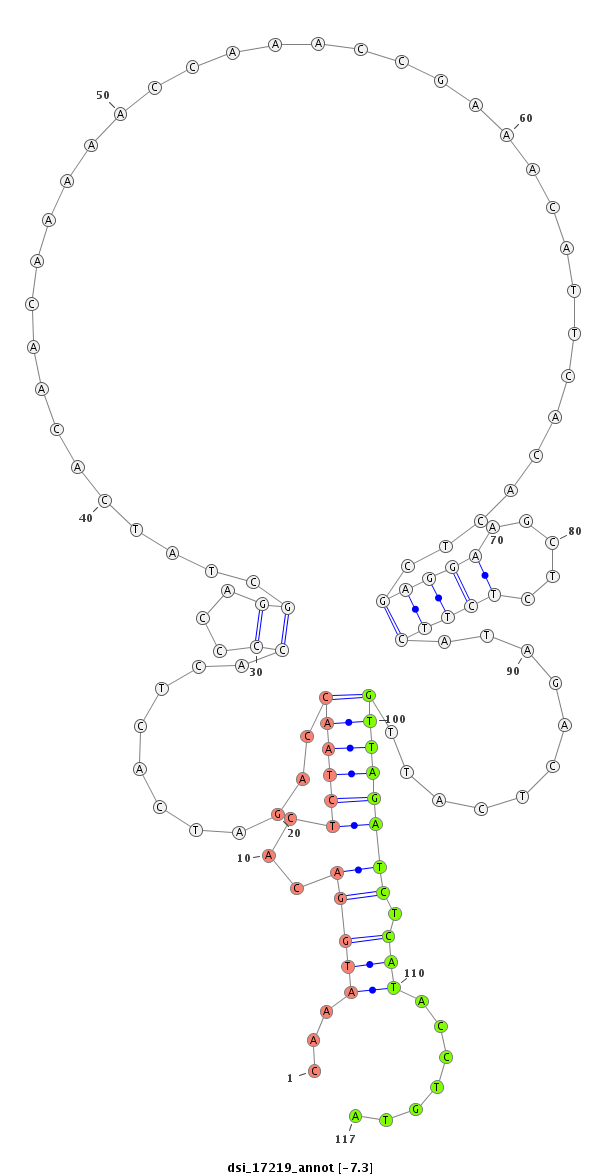
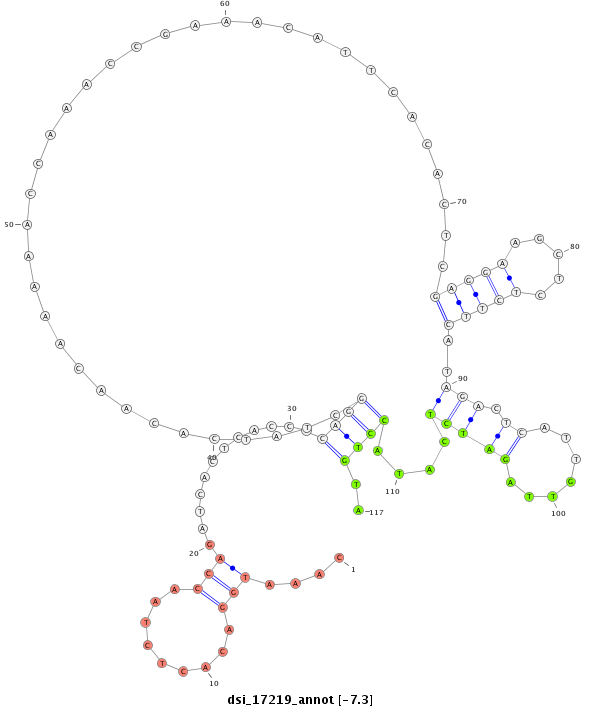
ID:dsi_17219 |
Coordinate:3l:15678936-15679085 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -7.3 | -7.3 | -7.2 | -7.2 |
 |
 |
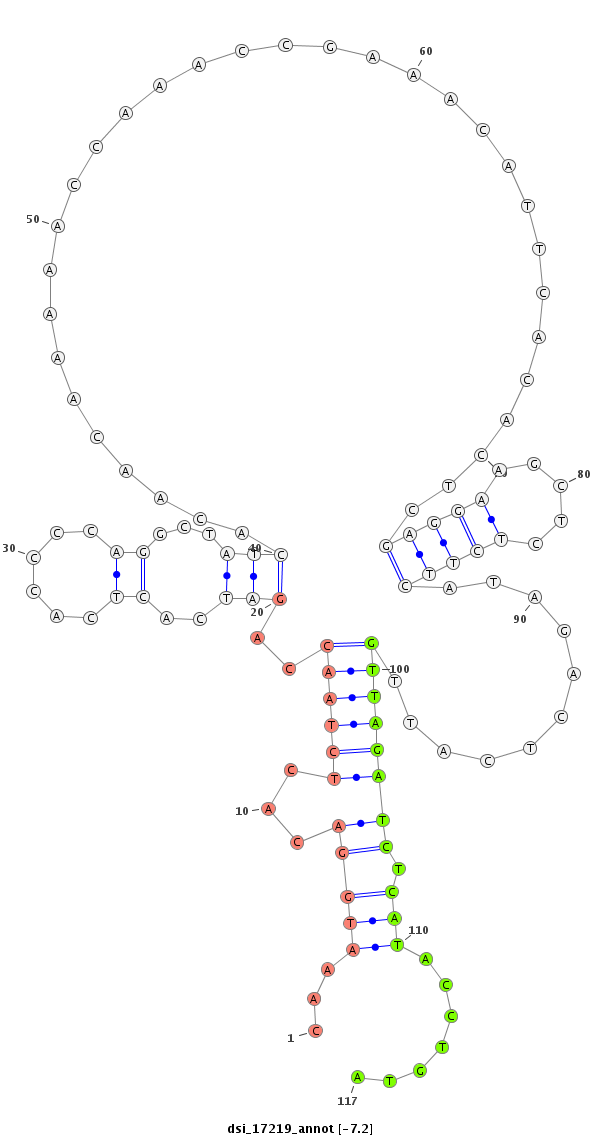 |
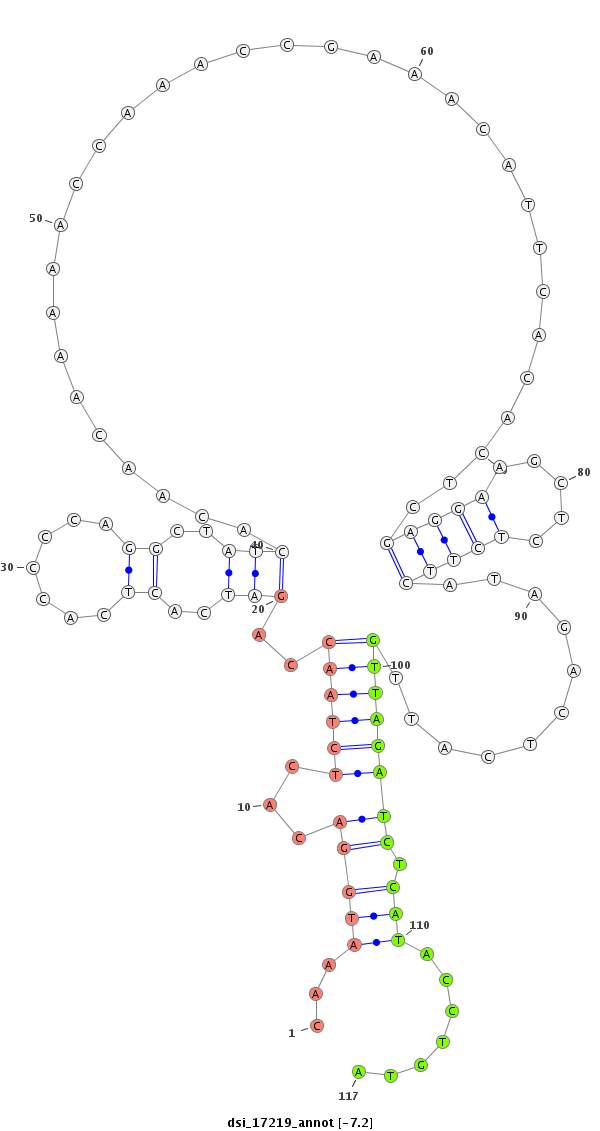 |
exon [3l_15678719_15678935_+]; CDS [3l_15678719_15678935_+]; intron [3l_15678936_15680198_+]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ATAACAACAATATCAGCACCACCACCACAAACACGATCAGCAACAGCGGCGTAAGTAGCCCAGTGTCCAAATGGACACTCTAACCAGATCACTCACCCCAGGCTATCACAACAAAAACCAAACCGAAACATTCACACTCGAGGAAGCTCTCTTCATAGACTCATTGTTAGATCTCATACCTGTAGTCCTAGCCGGGATTACAGGCCCCATCCACACACGATGTCCACATGCAGTTCACAACCACTCAAAC *******************************************************************....((............(((((...((....((.((((...........((....(((...........))).))...........)))).))...))...)))))........))****************************************************************** |
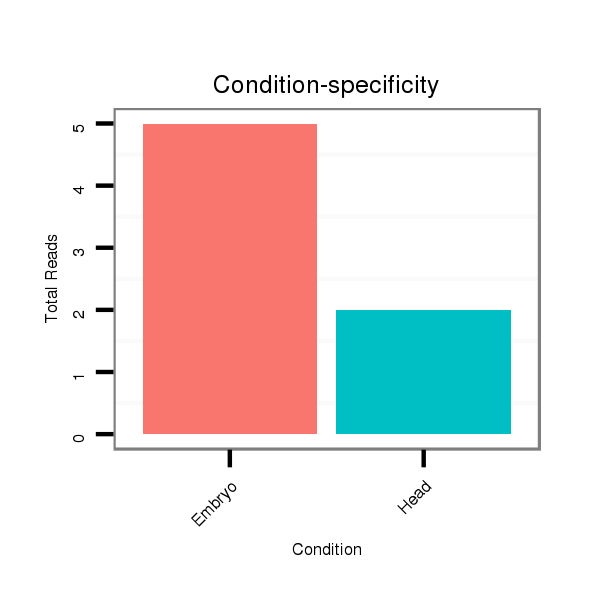
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
M023 head |
M024 male body |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR902009 testis |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................GTTAGATCTCATACCTGTA.................................................................. | 19 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................CAAATGGACACTCTAACCAG................................................................................................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................AAACACGATCAGCAACAGCGGC........................................................................................................................................................................................................ | 22 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...........................CAAACACGATCAGCAACAGCGGC........................................................................................................................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................CATTGTTAGATCTCATACCTGTA.................................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................ACGATCAGCAACAGCGGC........................................................................................................................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................CATGTCCACATGGAGTACACA........... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................TTTACTCCTAGCCGGG...................................................... | 16 | 2 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................GCGCATTGTTAGATTTCATA........................................................................ | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................CGTCATAGACTCTTTGT................................................................................... | 17 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................TCTAAACAGATCTCTCACCT........................................................................................................................................................ | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................ATTCCGGTAGTCCTA............................................................ | 15 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ....................................GCAGCAACAGCGGCGTA..................................................................................................................................................................................................... | 17 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................CCCGGTAGTCCTAGCC......................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................GCTCTAAACAGATCTCTCACC......................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................AGACTAAGTGTTAGATCTT........................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TATTGTTGTTATAGTCGTGGTGGTGGTGTTTGTGCTAGTCGTTGTCGCCGCATTCATCGGGTCACAGGTTTACCTGTGAGATTGGTCTAGTGAGTGGGGTCCGATAGTGTTGTTTTTGGTTTGGCTTTGTAAGTGTGAGCTCCTTCGAGAGAAGTATCTGAGTAACAATCTAGAGTATGGACATCAGGATCGGCCCTAATGTCCGGGGTAGGTGTGTGCTACAGGTGTACGTCAAGTGTTGGTGAGTTTG
******************************************************************....((............(((((...((....((.((((...........((....(((...........))).))...........)))).))...))...)))))........))******************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR1275487 Male larvae |
SRR553488 RT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................GGTGTTGGTGTTTGTTCTAGT................................................................................................................................................................................................................... | 21 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................GGTGGCGGTGTCTGTGCT...................................................................................................................................................................................................................... | 18 | 2 | 11 | 0.36 | 4 | 3 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................................................................................................................GGTGTTCGTCAGGTGTTCGTG...... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................CGCGTAACAATCTAAAGTAT........................................................................ | 20 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................TACAGGTTCACGACAAGTGT........... | 20 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................ATCCGAGTACCAATCT............................................................................... | 16 | 2 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................TTCGTAAGTCAGAGCTCCT.......................................................................................................... | 19 | 3 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................GAGCTCCTTAGAGACAA................................................................................................. | 17 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................GTTTGGCTATGAGATTGGT.................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
Generated: 04/23/2015 at 05:18 PM