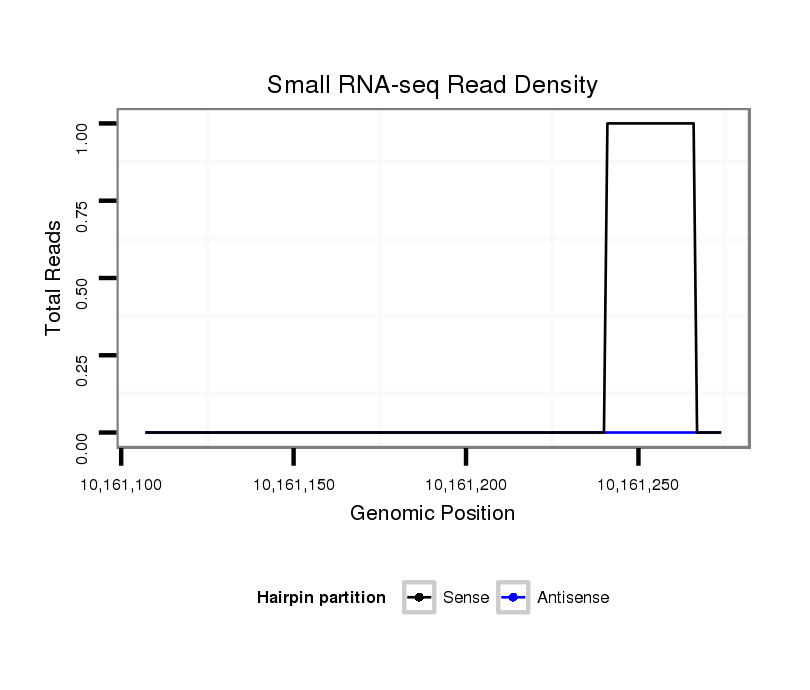
ID:dsi_17204 |
Coordinate:3r:10161157-10161224 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
five_prime_UTR [3r_10161141_10161156_+]; exon [3r_10161141_10161156_+]; five_prime_UTR [3r_10161225_10161243_+]; CDS [3r_10161244_10161444_+]; exon [3r_10161225_10161444_+]; intron [3r_10161157_10161224_+]
No Repeatable elements found
| ----------------------------------################--------------------------------------------------------------------################################################## GTTCTTATTGATCGTGGTATTTTTTGGCGCTCTGGCAGAGGTCACTCTAAGTAAAATTTGAGATATCATTTTATGTTTAGTTTAATAATTATTGGAATAGGACTACCAGCGACAACAGGTGATGCCGGGCAGCCAGGATGAGTGCACTTCTAAACGAAATCCAGCGGA **************************************************.......(((.................((((((................))))))..........)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR618934 dsim w501 ovaries |
SRR902009 testis |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................AGGATGAGTGCACTTCTAAACGAAAT........ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................ACAACAGGTGGTGCCGGGCA..................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....TATTGATCGGGGTTTTTTT................................................................................................................................................ | 19 | 2 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................ATAATTAGTAGAATAGGA.................................................................. | 18 | 2 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................GTCAGGATGAGTGCTCT.................... | 17 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....TATTGATCGTGGTAGTTTATA.............................................................................................................................................. | 21 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................CCAAGATGAGTGCTCTCCT................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................ATAATTATTGGATTAAGA.................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
|
CAAGAATAACTAGCACCATAAAAAACCGCGAGACCGTCTCCAGTGAGATTCATTTTAAACTCTATAGTAAAATACAAATCAAATTATTAATAACCTTATCCTGATGGTCGCTGTTGTCCACTACGGCCCGTCGGTCCTACTCACGTGAAGATTTGCTTTAGGTCGCCT
**************************************************.......(((.................((((((................))))))..........)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM343915 embryo |
SRR618934 dsim w501 ovaries |
SRR902008 ovaries |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................CCTCGGCCCTACTCACGTTAA................... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................GGTCGGGGTTGTCCACCACGG.......................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| .GAGAAAAACTAGCACAATAAAA................................................................................................................................................. | 22 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........ACTAGCAGCATAAAAAAA.............................................................................................................................................. | 18 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................AAAAACTGCAAGACCGTC.................................................................................................................................. | 18 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...............................TAGCGTCTCCAGTGAG......................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....AAAAACTAGCAGCCTAAAAA................................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
Generated: 04/24/2015 at 10:33 AM