
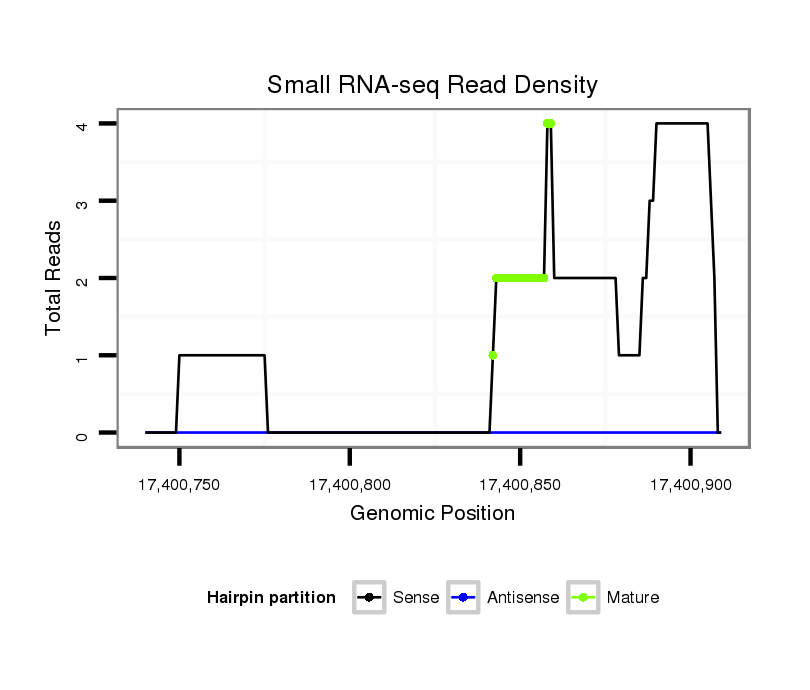

ID:dsi_16426 |
Coordinate:3r:17400790-17400859 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -9.5 | -9.4 | -9.4 |
 |
 |
 |
exon [3r_17400164_17400789_+]; CDS [3r_17400164_17400789_+]; CDS [3r_17400860_17401186_+]; exon [3r_17400860_17401296_+]; intron [3r_17400790_17400859_+]
No Repeatable elements found
| ##################################################----------------------------------------------------------------------################################################## TCAATGAGTTTGAGTTCTCCTGGCAGAAGATAGCTGATATTCAACTGCAGGTGGGTGCTTTTTCTTTCAGTTAAAATTGTAAGCTATGGATTAAGGTGATCTAATTCTTATCCGTTGCAGCTGGAGAAACTGATCGCCAAGAACTATTTCCTAAACCAGTCGGCCAAGGA **************************************************................................((.((((((.((((.........)))))))))).))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M024 male body |
M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................TCAACTGCAGCTGGAGAAACTGATCGCC................................ | 28 | 3 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................AGCTGGAGAAACTGATCGCCA............................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................AGAACTATTTCCTAAACCAGTCGGCCAA... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........TGAGTTCTCCTGGCAGAAGATAGCTG...................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TTTCCTAAACCAGTCGGCCA.... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................AATTCTTATCCGTTGCAG.................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................AACTGCAGCTGGAGAAACTGATCGCC................................ | 26 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................ATTCTTATCCGTTGCAG.................................................. | 17 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................TCCTAAACCAGTCGGCCAAG.. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................CTAAACCAGTCGGCCAAG.. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................ACTGCAGCTGGAGAAACTGATCGC................................. | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................TAGCTGATATTCAACTGCAGCTGGAG.................................................................................................................. | 26 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....TAAGTTTGAGTTTTTCTGGCAGAAGA............................................................................................................................................ | 26 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................GCTGGAGAAACTGATC................................... | 16 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TCAACTGCAGCTGGAGAAACTGA..................................... | 23 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................CAGCTGGAGCAACTGCTCGC................................. | 20 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................AATTGTAATCTATGGA................................................................................ | 16 | 1 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .TAATGAGATTGAGGTCTCC...................................................................................................................................................... | 19 | 3 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................TTGTAATCTATGGAGTA............................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................TTGTAGCTGGAGAAA......................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
AGTTACTCAAACTCAAGAGGACCGTCTTCTATCGACTATAAGTTGACGTCCACCCACGAAAAAGAAAGTCAATTTTAACATTCGATACCTAATTCCACTAGATTAAGAATAGGCAACGTCGACCTCTTTGACTAGCGGTTCTTGATAAAGGATTTGGTCAGCCGGTTCCT
**************************************************................................((.((((((.((((.........)))))))))).))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................AGCGGTTCTGGATAAAGGAAT................ | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........AAACTCACGAGGTCAGTCTT.............................................................................................................................................. | 20 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................TAGGCAACATCGAACTC............................................ | 17 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................TGGACAGCCGGTTCC. | 15 | 1 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................GACCGTCTCCTATCG........................................................................................................................................ | 15 | 1 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................GAATTTGGTGAGCCGGTT... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .................AGGAACGTCTCCTATCG........................................................................................................................................ | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
Generated: 04/23/2015 at 07:30 PM