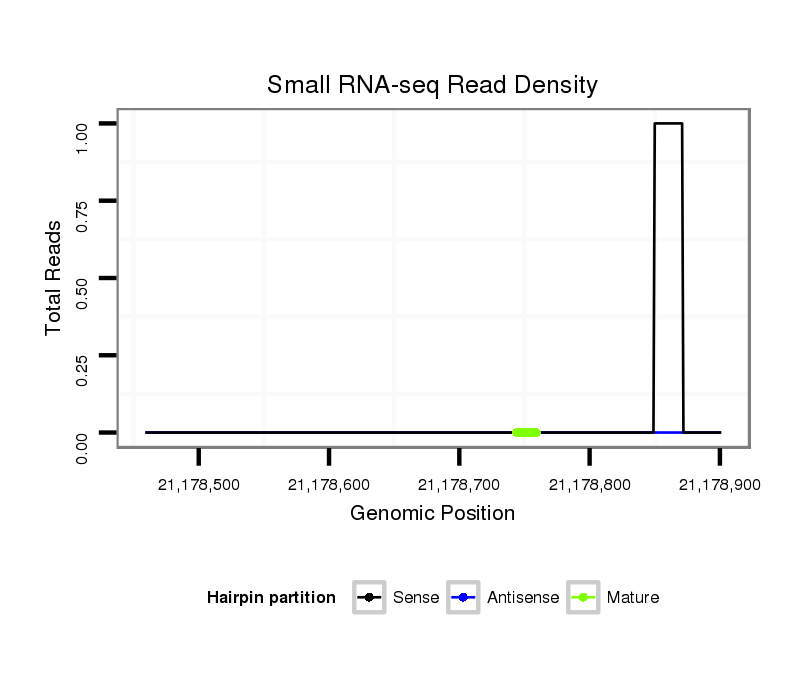
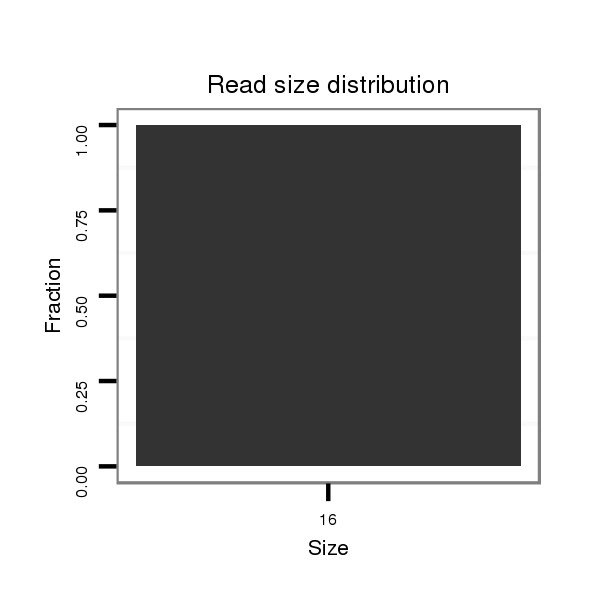
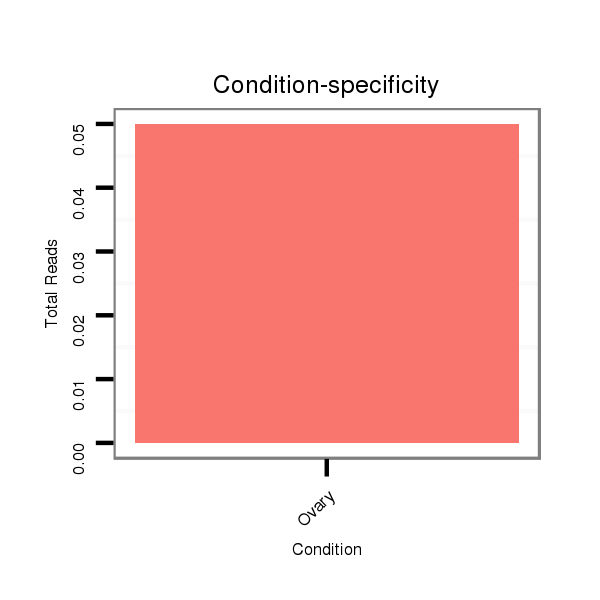
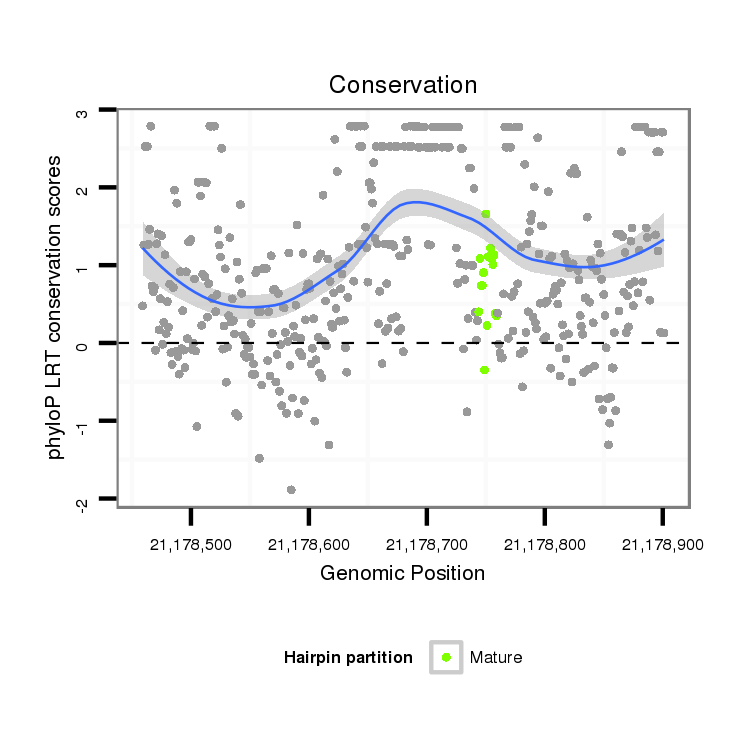
| droSim2 |
3r:21178459-21178901 + |
dsi_15783 |
GCGCGGTTTTTGCTGCTAT-CGAGT------------------------------TC---TTC-TTTTGCTGGA-----------------------------------TTTT-ATATATGGTAACGA-----------------------------------------------------------------------------------TTGTTGCTGCTGCT-GCTGCTGTATCATTGAAA-----------------------------------------------------------------------------------------------------------------A--------------------AT----------------------------------GCAGCTTTAAATTTAACTTAAATGCTA-AAACT------------TTGCA----------------AGTGGCTTTG--AAAA-------------GCAACGC------------------------------------------------GAGAGTT-------AGGACAA---------AC----AAGTTGGAGGAAA-----TTTTCCAGC-TACA------------GC---TGC---------------TGCCTC--------------------------TGCCAGCGTCGCAG-----------------------------------------------------------------TCGCTGTTCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTCCTCTTGT--T-------TCTTTT-T--------CCGCTGCTGCTGCTGC------------------------------------------------TACATTTGCTG------CTTGGC-ATTTT-CC--------------------------------------------------------------------------------------------GCA---C------------------------------------------AGCAACAACAATCGTT--------------------------------------------------------------------------------------------------------------------GAGACA---G-----------GACCGCAAGTTT----T-------------------GCT------AAAATGT----------------------ATCATAACACCAAATGAGCAG-------AGTTG-------GGAAGAGTGAGCGC-CA---AATAATGC-AA------ATAAATGAATAAGCC-----CAAA |
| droSec2 |
scaffold_13:460897-461132 + |
|
GCGCGGTTTTTGCTGCTAT-CGAGT------------------------------TC---TTC-TTTTGCTGGA-----------------------------------TTTT-ATATATGGTAACGA-----------------------------------------------------------------------------------TTGTTGCTGCTGCT-GCT---GTATCATTGAAA-----------------------------------------------------------------------------------------------------------------A--------------------AT----------------------------------GCAGCTTTAAATTTAACTTAAATGCTA-AAACT------------TTGCA----------------AGTGGCTTTA--AAAA-------------GCAACGC------------------------------------------------AAGAGTT-------AGGACAA---------AC----AAGTTGGAGGAAA-----TTTTCCAGC-TACAGCTAC------AGC---TGC---------------TGCCTC--------------------------TGCCAGCGTCGCAG-----------------------------------------------------------------TCGCAG-TCGAGATAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| dm3 |
chr3R:21681964-21682398 + |
|
GCGCGGTTTTTGCTGCTAT-CGAGT------------------------------TC---TTC-TTTTGCTGGA-----------------------------------TTTT-ATATATGGAAACGA-----------------------------------------------------------------------------------TTGTTGCTGCT----------GTATCATTGCAG-----------------------------------------------------------------------------------------------------------------C--------------------TT----------------------------------GCAGCTTTAAATTTAACTTAAATGCTA-AAACA------------TCG-----------------------TTTTG--AAAA-------------GCAACGC------------------------------------------------GAGAGTT--------AGACAA---------AC----AAGTTGGAGGAAA-----TTTTCCAGC-TACAGCTAC------AGC---TGC---------------TGCCTC--------------------------AGCCAGCGTCGCAG-----------------------------------------------------------------TCGCTGTTCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTCCTCTTGT--T-------TCTTTT-T--------CCGCTGCTGCTGCT---------CGTGC------------------------------------TACATTTGCTG------CTTGGC-ATTTT-CC--------------------------------------------------------------------------------------------GCA---C------------------------------------------AGCAACAACAATCGTT--------------------------------------------------------------------------------------------------------------------GAGACA---G-----------GACCGCAAGTTT----T-------------------GCT------AAAATGT----------------------AACATAACACCAAATAAGCAG-------AGTTA-------GGAAGAGTGAGCGC-CA---AATAATGC-AA------ATAAATGAATAAGCC-----CAAA |
| droEre2 |
scaffold_4820:6367085-6367537 - |
|
GCGCGGTTTTTGCTGCTAT-CGAGT------------------------------TCATTTTT-TTTTGCTGGA-----------------------------------TTTT-A--TATGGTAACGA-----------------------------------------------------------------------------------TTGTTGCTGCTGCT-------GTATCATTTA---------------------------------------------------------------------------------------------------------------TAAAA--------------------AT----------------------------------GTCACTTAAGATTTGATTTAATTGCTA-TCACA------------TCGCT----------------AGTGGCTTTGGCAAAA-------------GCAGCGC------------------------------------------------GACAGTT--------AGACAA---------AC----AAGTTGGAGGAAA-----TTTTCCAGC-TTCAGCTAC------AGC---TGC---------------TGCCTC--------------------------GGCCAGCGTCGCAG-----------------------------------------------------------------TCGCTGTTCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTCCTCTTGT--T-------TCTTTT-T--------CCGCTGCTGCTGCTGC------------------------------------------------TACAATCGCTG------CTTGGC-ATTTT-CC--------------------------------------------------------------------------------------------GCA---C------------------------------------------AGCAACAACAATCGTT--------------------------------------------------------------------------------------------------------------------GAGACA---G-----------GACCTCAAGTTT----A-------------------GCT------AAAATGT----------------------AACATAACACCAAATAAGAAG-------CGAAA-------AACAAAGTGAACGC-CA---AGTAATAC-AAATACTCGTATATGAATAAGCC-----CAAA |
| droYak3 |
3R:24799593-24800067 - |
|
GCGCGGTTTTTGCTGCTAT-CGAGT------------------------------TC---TTCTTTTTGCTGGA-----------------------------------TTTT-A--TATGGTAACGA-----------------------------------------------------------------------------------TTGTTGCTGCTGCT-------GTATCATTGAAAATCA----------------------------------------------------------------------------------------------------------------------------------TT----------------------------------GTCACTTCAAATTTGATTTAATTGCCC-AAACA------------TCGCA----------------AGTGTCAATG--AAAA-------------GCAATGC------------------------------------------------GAGAGTT--------AGACAA---------AC----AAGTTGAAGGAAA-----TTTTCCAGC-TACAGCTAC------AGC---TGC---------------TGCCTC--------------------------GGCCAGCGTCGCAG-----------------------------------------------------------------TCGCTGTTCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTCCTCTTGT--T-------TCTTTT-T--------CCGCTGCTGCTGCTGC------------------------TGCCACTGCTGCTGCGGCTGCTGCTACATTCGCTG------CTTGGC-ATTTT-CC--------------------------------------------------------------------------------------------GCA---C------------------------------------------AGCAACAACAATCGTT--------------------------------------------------------------------------------------------------------------------GAGACA---G-----------GACCTCAAGTTT----A-------------------GCT------AAAATGT----------------------AACATAACACCAAATAAGAAGCGAACAGAAC--AC-----AAAAAAGTGAGCGC-CA---AATAATGC-AA------ATAAATGAATAAGCC-----CAAA |
| droEug1 |
scf7180000409682:944865-945360 - |
|
GCGCGGTTTTTGCTGCTCT-CGAGTTTTT---TT---------------------TC---TTC-TTTTTCTGGA-----------------------------------TTTC-A--CATGGTAACGA-----------------------------------------------------------------------------------TTGTTGCTGCTGCT-------GTCTAATTGAAA----------------------CCT----------------------------------------------------------TAAAAGTTCTTATTTTA-TTTTTAAA----------------------------------------------------CTA--------------------------------TAAATTA------------TCACT----------------AGTCCCTTAG--AAAA-------------GTAGC-C---AAAAA--------------------------------------ATGAGAATT--------CGACAA---------AC----AAGTTGGAGGAAA-----TTTTCCAGC-TACAGCTAC------AGCTGCTGC---------------TGCCTC--------------------------TGCCAGCGTCGCTG-----------------------------------------------------------------TCGCTGTTCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTCTTCTTGT--T-------TCTTTTTT--------CCGTTGCTGCTGCTGC------------------------TGCCTTTGC---------------T---TCTGCTGAATTTTCATGAC-ATTTT-CC--------------------------------------------------------------------------------------------ATACAGAAA-----------AC----------------AA---------AGCAACAACAATCGTT--------------------------------------------------------------------------------------------------------------------GAGACA---GG----------GACCTTAAGTTT----TTCGATATA-----------TCTAGT---TAAATGTAATAAACAAAGAAAGAAAAAGTTAAATAA-----------------------A--AT-------AAAAGTGAGCGC-CA---AATTATGG-AA------ATAAATAAATAAGCC-----GAAA |
| droBia1 |
scf7180000302113:1863689-1864132 + |
|
GCGCGGTTTTTCTAGCTCT-CGAGC------------------------------ATTTTTTT-TATAACTAGA-----------------------------------TTCC-G--CATGCTAACGA-----------------------------------------------------------------------------------TTGTTGCTTCT-------------------------------------------------------------TAA----------------------------------------------------------------------------------------------AAAAATTAT-----AATTTT-TTGAGTAA-------------------------------TAAA-CAA-----------TCCTAAAA----------------ATTGTTG--AAAA-------------GTATC-T------------------------------------------------A--------------AGAC------------C---------AGGGGAAA-----TTTTCCAGC-TAAGGCTTAGGCTACAGC---TGC---------------TGCCTC--------------------------TGCCAGCGTCGCTGCCGCAGCC----------------------------------------------GCTGTCGCTG-TCGCTGTTTGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTCTTCTTGT--T-------CCTTTTTT--------CCGCTGCTGCTGCTG---------------------------------------------------------CTG------CCTGGCCATTTT-CCTTTGGC-------------------------------------------------------------------------------------------------------------------------------------------ACAACAAGCGTT--------------------------------------------------------------------------------------------------------------------GAGAAA---G-----------GACCTCAGCTT-GATGTGCTATATACTTGCCTTATAGCT------AAAATGT----------------------AATAAA--ATCGA----GCA-------AAGC--AA----ATATTAAGTGAGCGG-GA---AAGTATGCGAA------ATAACTGAATAAGCC-----GAAA |
| droTak1 |
scf7180000415658:114849-115283 + |
|
GCGCGGTTTTTCCAGCTCT-CGAGT------------------------------AAATTTTT-TTAATCTAGC-----------------------------------TTTTGA--AATGTTAACGA-----------------------------------------------------------------------------------TTGTTGCTGCTCCA-GCTGCTA------------------------------------------------------------------------A--------------------------------------------------------------------------AAATAATATTTGAAAATTTT-AAACGTA--------------------------------TAAAATA------------TCGTA----------------AATATTTTCG--AAAA-------------CTAAC--------------------------------------------------AAGA----------------------------------GTCAGGGGAAA-----ATTTCCAGC-TAAAGCTACAGCTACAGC---TGC---------------TGCCTC--------------------------TGCCAGCGTCGCTGCCGCC----------------------------------------------------G-------TCGCTGTTTGAGATAAAGGATATGCTCGGATCTAAAAGATACAGTTTTTCTTCTTGT--T-------TCTTTT-T--------CCGCTGCTACTGCTGC------------------------------------------------T------GCTG------CTTGGG-ATTTT-CC--------------------------------------------------------------------------------------------CTA---C------------------------------------------AGCAACAACAATCGTT--------------------------------------------------------------------------------------------------------------------GAGACA---G-----------GACCTTTAGTATTATTTGCTATATATTTGCTTTATGGCC------AAAATGT----------------------AATAT----CCAA-----------------------------AAAAGTGAGCGG-GA---AAGTATGCGAA------ATAACTGAATAAGCC-----GAAA |
| droEle1 |
scf7180000491261:1068543-1068950 - |
|
GCGCGGTTTTTGCTGCTCC-CGAGTTT---------------------------------TT----ATTCTGGA-----------------------------------TTTC-A---ATGGTAACGA-----------------------------------------------------------------------------------TTGTTGCTGCTGCT-------GTATTGTCAAGG----------------------ACTATT---ATATTACCCAT--------------------------AAAAAATAATTTAT-------------------------------------------------------------------------------------------------------------------------------------TCCAA----------------AGTATTTTTG--AAAA-------------ATAAT-C------------------------------------------------AAAAATT--------GGACAA---------AC----AAGTTGGAGGAAA-----TTTTCCAGC-TACAGCTAC------AGC---TGC---------------TGCCTC--------------------------TGCCAGCGTCGCTG-----------------------------------------------------------------CCGCAGTTCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTCTTCTTGT--T-------TCTTTTTTT-------CTGCTGGTGCTGCTGC------------------------------------------------T------TCTG------CTTGGC-ATTTT-GC--------------------------------------------------------------------------------------------ACA--GCAAC---------------------------------------AGCAACAACAATCGTT--------------------------------------------------------------------------------------------------------------------GAGAAA---G-----------GACCACAAGTTT----T-------------------GCTAGAT--AAAATGT----------------------AACATACAACCAA--------------------------GAAATAAGTGAGCGC-CA---AAGTATGC-AA------ATAA-------------------- |
| droRho1 |
scf7180000761346:13566-13977 + |
|
GCGCGGTTTTTGCTGCTCC-CGAGTTT---------------------------------TT----TTTCTGGA-----------------------------------TTTC-A---ATGGTAACGA-----------------------------------------------------------------------------------TTGTTGCTGCTGCT-------GTTTCGTTGGGA----------------------ACTAGTCAAATATTACTTAT--------------------------CAAAA-------------------------------------------------------------------------------------------------------------------------ATA-AAATA------------TT--C----------------AGTATTTTTG--AAAA-------------GTAAC-CAAC----------------------------------------------AAAATT--------GGACAA---------AC----AAGTTGGCAGAAA-----GTTTCCAGC-TACA------------GC---TGC---------------TGTTTC--------------------------TGCCAGCGTCGCTG-----------------------------------------------------------------CCGCTGTTCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTCTTCTTGT--T-------CCTTTTTT--------GCGCTGCTGCGGCTGC------------------------------------------------T------TCTG------CCTGGC-ATTTT-CC--------------------------------------------------------------------------------------------ACA--GCAAT---A------GC---------------------------AACAACAACAATCGTT--------------------------------------------------------------------------------------------------------------------GAGACA---G-----------GACCTTAA----------------------------GCT------AAAATGT----------------------AACTTACAACCAA-T-------------------------AAAGAAGTGAGCGC-CA---AAGTATGC-AA------ATAAATGAATAAGCC-----GAAA |
| droFic1 |
scf7180000454055:1583337-1583777 + |
|
GCGCGGTTTTTGCTGCTCC-CGAGTTT---------------------------------------TTTCTGGA-----------------------------------TGT--A--AATGGTAACGA-----------------------------------------------------------------------------------TTGTTGCTGCTGCT-------GCTTTATTGAAA----------------------ACTAA-------------------------------------------------------------------------------------------------------------AATTAATATT----AATT---------------------------------TTAAAAGTTT-AAACAACCCAGAGCTTGTC-AA----------------AGTTTTTTCG--AAAA-------------GTATA-C------------------------------------------------AAAAATC--------GGACAA---------AC----AAGTTGGAGGAAA-----TTTTCCAGC-TACA------------GC---TGC---------------TGCCTC--------------------------TGCCAGCGTCGCTG-----------------------------------------------------------------CCACTGTTCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTCTTCTTGT--T-------CCTTTT-T--------CCGCTGCCGCTGCTGCTGCTGCTCGTGC------------------------------------T---TCTGCTG------CCTGGC-ATTTT-CC--------------------------------------------------------------------------------------------ACA--GC------------------------------------------AGCAACAACAATCGTT--------------------------------------------------------------------------------------------------------------------GAGACA---G-----------GACCTTAGGTAT----T-------------------GCTATGTCAAAAATGT----------------------AATATACAACCGA--------------------------AAATTAAGTGAGCGC-CA---AAGTATGC-AA------ATAAATGAATAAGCC-----GAAA |
| droKik1 |
scf7180000302475:2818732-2819221 - |
|
GCGCGGTTTTGGCTGCTCCCCGAGTTT---------------------------------TT--TTGTGCCTGA-----------------------------------TTTT-T--TATGGTAGCGA-----------------------------------------------------------------------------------TTGTTGCTGCT-CTGACAGCT------------------------------------------------------------------------------------------------------------------------CAAGAAAATTGCCACGCAGAAAAAAAAG-------AA-----AATATTGCCGAATAAAAATTG--------------------------------------------TCACAAAAGTTGTT-------GG-----------------------------------CCAAAAAGAAA------------AAAAGATTTGTGTTAGCCGAAA-----AGGC--------AGCCAA---------AC----AAGTTGGAGGAAA-----TTTTCCAGC-TACA------------GC---TGC---------------TGCCGC--------------------------AGCCAACGTCGCTGTCGCA----------------------------------------------GC-GCTGCCGCTGT-----GTCCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTCTT-------T-------GCTTTT-T---------CG-----------------------------CAGG------------------------------------CTG------CTTGGC-ATTTT-CC--------------------------------------------------------------------------------------------ACA--GC------------------------------------------GGCAACAACAATCGCCAACCAAAGAAG-AATTTCGTCTGCCAGCCGAAAAAAAAGAA---GAAA-T------------AAT---------------------------------------------------AA---------------------------------------------------------G------AAAATGA----------------------AACATAAAGCCAG-TGAGT------------GAGAGAGAGAGAGAAGAGAACGC-CA----AGAATGC-AAAA----ATAAATGAATAAGACCCCCAGAAA |
| droAna3 |
scaffold_13340:11464459-11464998 - |
|
GCGCGGTTTCTGCTGATAC-CGAATC------------------------------------------TCTAGA-----------------------------------ATT-----AATGGTAACGA-----------------------------------------------------------------------------------TTGTTGCTGCTGCT-GCTGTTG----A-----------------------------------------------------------------------TTT-------------TTTGAGAGT---------ATTTTTTAAAAGAAAAATTAGCTTACAATAAAAAAAA-------TA-----AATA------AATAAATA--G--------------------------------------------CC----------------------------------------------------------ATAGAGAAATGATCGAGCGGCAAAAGGTATATATTTTTCGAAAGG---A--AAACCCCAAG---GA---------AC----AAGTTGTGGGAAA-----TTTTCCAGC-TACGGCTAC------AGC---TGC---TGCTGTCGGTGCTGCCTC--------------------------AGTCAACGGCTCTGCAGCA----------------------------------------------GC-GCTGTCGCTGT-----GTCCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTCTTCTTTT--T-------CCTTTTT----------TGCTGCTGC------------TTGAGTTGTGCAGG------------------------------------CTG------CTTGGT-ATTTT-TCTTTATCCCA-------------------------------------------------------ACAGTAACAGCTAAGAAAAAAAA-----------AAAA---A------ACAAAAACAACTACG---------GCAACAGCAAGAACAATCGGT---------------------------------------------------------ATTTTAACGTGAAATTAACGAAGGAATT-------------------------------TAGCCA-----------------------------------------------GGTAGCA------AAAATGT----------------------A------------------------------------------------AGCAC-CGAGGGAGTATGC-AA------ATAAATTAATAAGCC-----AAAG |
| droBip1 |
scf7180000396691:373322-373837 - |
|
GCGCGGTTTCTGCTGATAC-CGAATC------------------------------------------TCCAGA-----------------------------------ATT-----AATGGTAACGA-----------------------------------------------------------------------------------TTGTTGCTGCTGCT-GCTGTTG----A-----------------------------------------------------------------------TTT-------------TT--------------------TTT--AAGAAAAATTAGCTTACAAAAA----AA-----TTAA-----AATA------AATAAATA--G--------------------------------------------TCACA---------------------------------------------------------------ATGATCGAGCGGCATTAAGTATATATTTTTTGAAAGT---A--AAACCCCAAG---GA---------AC----AAGTTGTGGGAAA-----TTTTCCAGC-TACGGCTAC------AGC---TGC---TGCTGTCGCTGCTGCCTC--------------------------TGTCAGCGCCTCTGCGGCA----------------------------------------------GC-GCTGTCGCTGT-----GTCCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTCTTCTTTT--T-------CCTTTTT----------TGCTTCTTC------------TTGAGTTGTGCAGG------------------------------------CTG------CTTGGT-ATTTT-TTTTT-----------------------------------------------------TAATCCAAACAGCAACAG------------------------AAAT---A------ACAAAAACAAGAACG---------GCAACA------GCA------------------A----------------------ACAGCA---AGAA-CAATCGGTATTTTAACGTGAAACTAACGAAGGAATT-------------------------------TAGCCA-----------------------------------------------GCTAGCA------AA----------------------------------------------------------------------AATGTGAGCAC-CGAGCGAGTATGC-AA------ATAAATTAATAAGCC-----AAAG |
| dp5 |
2:23406550-23407102 - |
|
GCGCGGTTTTTGCTGCTCT-CCAGCTT-----------------------------------------GGGGGGATTTGTGGTAAAACTTCTATCTT----------------------------CTG-----------------------------------------------------------------------------------TTGTTGCTG------------------------------------------------------------------GGTCCTCGTCCTCCTGTTGTTAAT--CACAGC------------------------------------------------------------------------AGAAA-----------------------------------------------------------------------------------------TTGTTG--AAACGAATTAGCGAGAAAGC----ACCAAAAATAAA------------A---------------CCGAGT-----AGGAAG--CCAAGGAAAA---------GC----A-----AAGGAAA-----TTTTCCAGC-TACAGCTAC------AGC---TGCTC-TGCTGCTACTGCTGCCGC--------------------------TGCGAGCGTCGCTGC----------GTCAACAGCGCTGCTGCTGCCGCGGCTGCTGCTGCT-GCTGCGGGTGCTGCTGT-----GTCCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTCTTCTTCTGTTTCAATTTTTTTTT-TTCGTGTTT--TTTGCTCC------------TTGTCTTGTGCAGG------------------------------------CTG------CTTGGC-TTTCTGGCTGCG---------------------------------------------------------------------------GAAAAGCAGAGCGGAA---C---AGC---------AA------CAACA---------GCAACA------GAAAT---------------A-----------------------------------------------------------CTAAAGCAATAATTTGTTTGGAGAATTCGTT--------------CCGACG-----------------------------------------------GCTGGCC-----AAAAATGT----------------------GGCATACAACT--------------------------------GAAGTGTGGCCCGAAGAAACTATGC-AA------ATAAATTAATATGCC-----GAAA |
| droPer2 |
scaffold_3:6178270-6178815 - |
|
GCGCGGTTCTTGCTCCTCT-CCAGTTT-----------------------------------------GGGGGA-TTTGTGGTAAAACTTCTATCTT----------------------------CTG-----------------------------------------------------------------------------------TTGTTGCTG------------------------------------------------------------------GGTCCTCGTCCTCCTGTTGTTAAT--CACAGC------------------------------------------------------------------------AGAAA-----------------------------------------------------------------------------------------TTGTTG--AAACGAATTAGCGAGAAAGC----ACCAAAAATAAA------------A---------------CCGAGG-----AGGAAG--CCAAGGAAAA---------GC----A-----AAGGAAA-----TTTTCCAGC-TACAGCTAC------AGC---TGCTC-TGCTGCTACTGCTGCCGC--------------------------TGCGAGCGTCGCTGC----------GTCAACAGCGCTGCTGCTGCCG------CTGCTGCT-GCTGCGGGTGCTGCTGT-----GTCCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTCTTCTTCTGTTTCAATTTTTTTTT-TTCGTGTTT--TTTGCTCC------------TTGTCTTGTGCAGG------------------------------------CTG------CTTGGC-TTTCTTGCTGCG---------------------------------------------------------------------------GAAAAGCAGAGCGGAA---C---AGC---------AA------CAACA---------GCAACA------GAAAT---------------A-----------------------------------------------------------CTAAAGCAATAATTTGTTTGGAGAATTCGTT--------------CCGACG-----------------------------------------------GCTGGCC-----AAAAATGT----------------------GGCATACAACT--------------------------------GAAGTGTGGCCCGAAGAAACTATGC-AA------ATAAATTAATATGCC-----GAAA |
| droWil2 |
scf2_1100000004943:14359177-14359742 - |
|
GCGCGGTTTTGCCTGCTTC-TG-CT------------------------------GC---TTC-TCTTGAACGG-----------------------------------TTTT-ATTTATGGTAACTAAAAAAAAAATAAAAAAAATTGAAAAATTAAACGTAAAAACCCAAACGAAATTCAATTAAATGTAATCACAACAAATTAATAAATTATTCAAGTTTG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGCTAAAAATTGTT-------TGTCACCTTTC---------------TAAATC----ACAAAAAATAA-----------------------------------------GGAAG--CAA-AAAAAAGAAAGAAAAACGAAACCGTTGAAGGAAA-----TTTTCCAGTTTACAGCTGC------TGC---------------------TGCTGC--------------------------TGCTGGTGTTGCTGCTGGC------GTC---------------------------------AACAGC-GCTGCTTGTAT-----GTCCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTCCTTTTTG--C-------CATTCATT--------CTCCTGCT---------------CGCCTCTTCC-------TGTC---------------------------CCTG------CTTGTG-CTTCT-CCTTCACCACCTTAGATGGCACTCCAGTTTACCTCAATAAACCAATTCACTTATTTTCTATCTCGGACAGCAGCAGC----------A-----ACAA---C---ATC---------AA------CAACA---------GCAACA------ACAAC---------------A-----------------------------------------------------------ATAA----------------------------------------------------------------------------------------------------------------------------------------------TAAC-AAATAA---------------------AGGTAGAAATGTGCTT-C----AACTATGC-AA------ATAAATTAATAAGCC-----AAAA |
| droVir3 |
scaffold_13047:16101865-16102392 - |
|
TTGCGGTTTGTTCTTAATT-TTCATTTATGATAT-------------------------------GCTGCT--------------------------------------TTTT-T-----------AA-----------------------------------------------------------------------------------TTGTTCTTGT-----CG--TCG----ATTGTTGATTGTTGTTCTT------------------------------------------GATGTTGTTA--------------------------------------------------------------------------------------------------------------------------CTA---------AA-ATG-----------TTCTAAAAATGAG-CG-TTGA---ATTTTCG--CAAA-------------GCAGCGT-------------------------------------------------------------------AA---------------------ACGAAAAT----TTTTCCAGC-TACAGCTG---------------CTGGCGGTGCCACGGCCGCAGCTGCTAACGCTGCTGCTGCT-CTGCTCTGCTAACGCCGCTGCTGCTGCTGCTG------------------------------CTCCTGACTGC-GCTGCTGCGTT-----GTCCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTTTTTTTA-----------------------------------------------------CTTGTGCAGGCAGC--------------------------------TTG------CTTGGC-TTTTT-GCTTTGACGCTGC-GTTTG------------------TAAGC----------------GAACTTAAGCAAC-------------------------A--ACAAC---A------ACAACAACAACAACG---------GCAAAA------GAA------------------A--------------------------------------------------------ATTTA-------------------TACTGTG---TGTGCGTGCGGCTTG-AGAGCAG-AATGA-TGTGAGTGACAA------------------------GTTGGCG------AAAAAAA---------------AAAA-----------ACCGA-----------AAACGAA--AC-------AAAAATGTGCGC------AACTATGC-AA------ATAAATTAATAAGCC-----AAAG |
| droMoj3 |
scaffold_6540:14905473-14906043 - |
|
GCGCGACTGGTGCTCAACT-TGTATTT---------------------------------ATC-TGCTGCTGC------------------------------------TTT--------------AA-----------------------------------------------------------------------------------TTGTTGTTGTTGCT-GCTGTCA----ATTGTTGATTGTGGTTCTTGTTGTTGTTG--------------------CGCCATTGGACTCTATTTGTCAA-------------------------------------------------------------------------------------------------------------------ATTTG------------------------------TCA-AAAAACCCG-CCTTTCA---TTTTTGG--CGTA-------------GCAGCGT-------------------------------------------------------------------AA---------------------ATGAAAATT---TTTTTCAGC-TACAGCTG---------------CTGGCGGTGCCACGGCCGCAGCTGCTAGTGCTGCTG------CTACTCTGCCAACGTCGCAGCCGCTGCT---TTC---------------------------------GGCTGC-GCTGCTGCGCT-----GTCCGAGATAAAGGATATGCCCGGATCTAAAAGATACAGTTTTTTTTTTT-----------------------------CGCC-------------------AACTTGTGCAGGCTGC--------------------------------CTG------CTTGGC-TTTTT-GCTGTGTCGCTTC-GTTCC------------------TAAGC----------------GAACTTAAGCAGTAACAG------------------------CAAC---AACAACCACAAC------AACAATCACA---GCAAC------AACATT----------------A----------------------ACAGCAGCAGCAAAG------------A-GAGAAAATTA-------------------CAATGTG---TGTGCGTGCGGTTTGGATAGCAG-AGTAAAAATGAGTGACAA------------------------GTTGGC-----------------------------AAAA-----------ACCAA-----------AAATG-A--AC-------AAAAGTGTGCGC------AACTATGC-AA------ATAAATTAATAAGCC-----AAAC |
| droGri2 |
scaffold_15116:946858-947431 - |
|
CCGCGGTTTGTTCTTAACT-TTTATTTATGGTATGCTGCTTTTGCTGCTGCTGCTG--------------------------------------CTACTGCTGCTGCTGTTTT-T-----------TT-----------------------------------------------------------------------------------TTCTTCTTGTTTGT-CG--TCG----ATTGTTGATTGT----------------------------------TAT--------------------------CAATT-----------------------------------CAAAAG--------------------AA----------------------------------TTCACT-------------------------------------------AAATGCGAG-CG-TTAA---ATTTT-------------------------------------------------------------------------------------------CGCCTA---------GC----AGCGTAGTCAAAATTTTTTTTTCCAGC-TACAGCTG---------------CTGGCGGTGCCACAGCCGCAGCTGCTCATGCTGCTTCTGCTGCTGCTCCGCTTACGTCGCAGCTGCTGCTGCTGCC---------------------------------GACTGC-GCTGCTGCGTT-----GTCCGAGATAAAGGATATGCCCAGATCTAAAAGATACAGTTTTTTTTT-------------------------------------------------------ACTTCTGCAGGCTGC--------------------------------TTG------CTTGGC-TTTTT-GCTGTGTCGCTCC-ATTCG------------------CAAGC----------------GAATTTAGGCAAC----------------G-----ACAA---G------A------ACAAC------AACAGCCAGAAGAACAACA------AA-------------------A----------------------ACAGCA---GAAAAC------------AACGTAAAATTA-------------------GAATGCGTGGTGTGCGTGCGGGTTG-AGAGTAA-AGTGA-AGTGAGTGACAA------------------------GTTGGC-----------------------------AAAA-----------ACTGA-----------AAATGTA--AC-------AAAAATGTGCGC------AACTATAC-AA---CAAGTAAATTAATAAGCC-----AAAG |