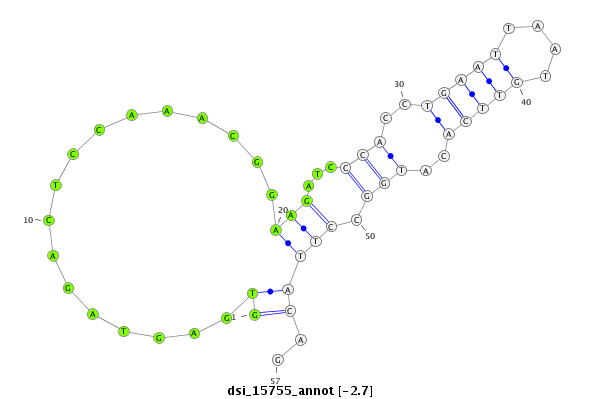
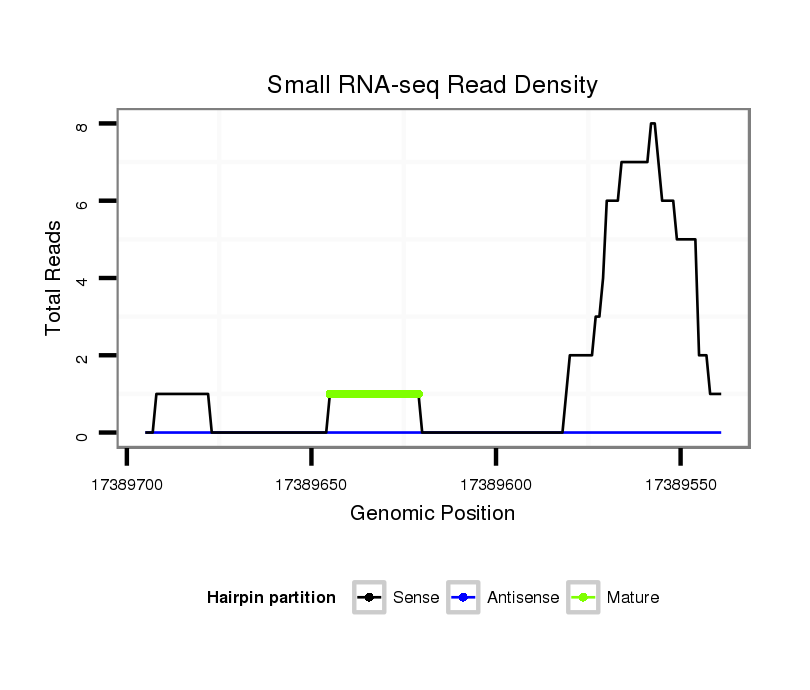
ID:dsi_15755 |
Coordinate:3r:17389589-17389645 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
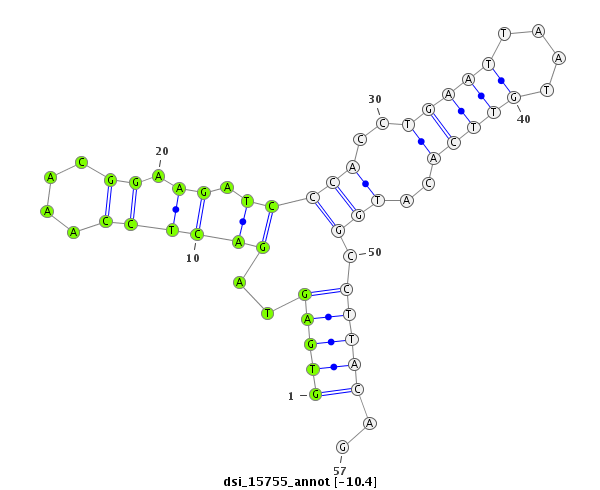
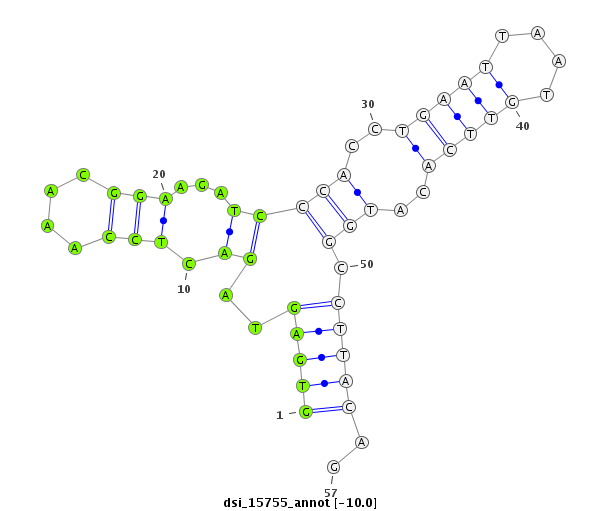
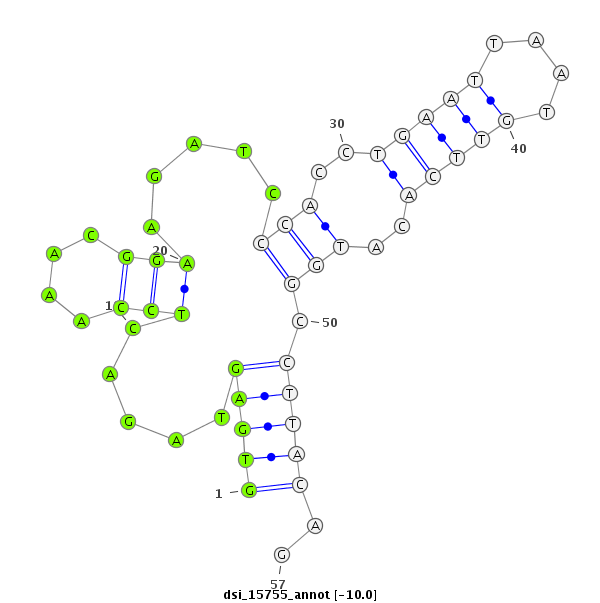
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -10.4 | -10.0 | -10.0 | -9.6 |
 |
 |
 |
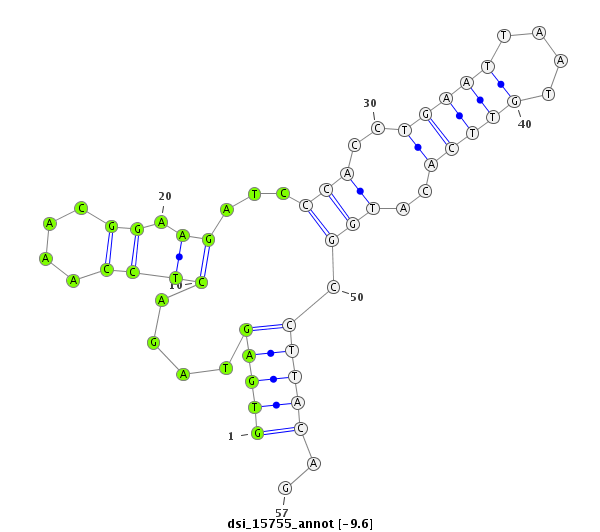 |
CDS [3r_17389333_17389588_-]; exon [3r_17389176_17389588_-]; CDS [3r_17389646_17390235_-]; exon [3r_17389646_17390264_-]; intron [3r_17389589_17389645_-]
No Repeatable elements found
| ##################################################---------------------------------------------------------################################################## GGCAAGGCCAAGGACTTCAGTCCGCGCGCTCGATTCCGCAGCTGGCTGGGGTGAGTAGACTCCAAACGGAAGATCCCACCTGAATTAATGTTCACATGGCCTTACAGATACGAGCTGCCATTCGATAGGCATGACTGGATAGTCGATCGCTGTGGCA **************************************************((.................(((...(((..(((((....)))))..))).)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR902008 ovaries |
SRR902009 testis |
SRR553486 Makindu_3 day-old ovaries |
M024 male body |
M025 embryo |
M053 female body |
SRR553485 Chicharo_3 day-old ovaries |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................TAGGCATGACTGGATAGTCGATCGC....... | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................CGCGCTCGATTCCGCAGCTGGCTGGGA.......................................................................................................... | 27 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTGAGTAGACTCCAAACGGAAGATC.................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................ATGACTGGATAGTCGATCGCG...... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................TGGGATACGAGCTGCCATTCGATAGGC........................... | 27 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................CATGACTGGATAGTCGATCGCTGT.... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................CGATAGGCATGACTGGAT................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................GTGAGTAGACTCCAAACGGAAA..................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................CTGCCATTCGATAGGCATGACTGGA.................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...AAGGCCAAGGACTTC........................................................................................................................................... | 15 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................GATAGTCGATCGCTGTGGCA | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................ATAGGCATGACTGGATAGTCGATCGC....... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................TGCCATTCGATAGGCATGACTGGATAGTC............. | 29 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................TTACGAGCTGCGATCCGATA.............................. | 20 | 3 | 8 | 0.88 | 7 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................GCAGCTGGCTGGGGT......................................................................................................... | 15 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................TGGCTGGGGTGAGTA.................................................................................................... | 15 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................GATTAATGTTCTCATGGACT....................................................... | 20 | 3 | 10 | 0.20 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................GGATAGTCGGTCGCT...... | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................................GTCGATCGCTCTGGC. | 15 | 1 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................TTGAGTAGACTCCAAAAGAAA...................................................................................... | 21 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CCGTTCCGGTTCCTGAAGTCAGGCGCGCGAGCTAAGGCGTCGACCGACCCCACTCATCTGAGGTTTGCCTTCTAGGGTGGACTTAATTACAAGTGTACCGGAATGTCTATGCTCGACGGTAAGCTATCCGTACTGACCTATCAGCTAGCGACACCGT
**************************************************((.................(((...(((..(((((....)))))..))).)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|
| .............................................................GGTTTGCCTCCTTGGG................................................................................ | 16 | 2 | 20 | 3.95 | 79 | 79 | 0 | 0 |
| ...............CAGTCAGGCGCGCGA............................................................................................................................... | 15 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ..........................................ACCGACCCCACTCAT.................................................................................................... | 15 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| ..............GAAGGCAGGAGCTCGAGCTA........................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 |
| ...................................................................................TAATTACAAGAGTACCTGAC...................................................... | 20 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 |
| .........................................................................................................................AGTTCCCCGTACTGACCTA................. | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 1 |
| ..................................................................................TTGATGACAAGTGTACC.......................................................... | 17 | 2 | 18 | 0.06 | 1 | 1 | 0 | 0 |
| ............................GAGTTTAGGCGTCGACCAA.............................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
Generated: 04/23/2015 at 06:35 PM