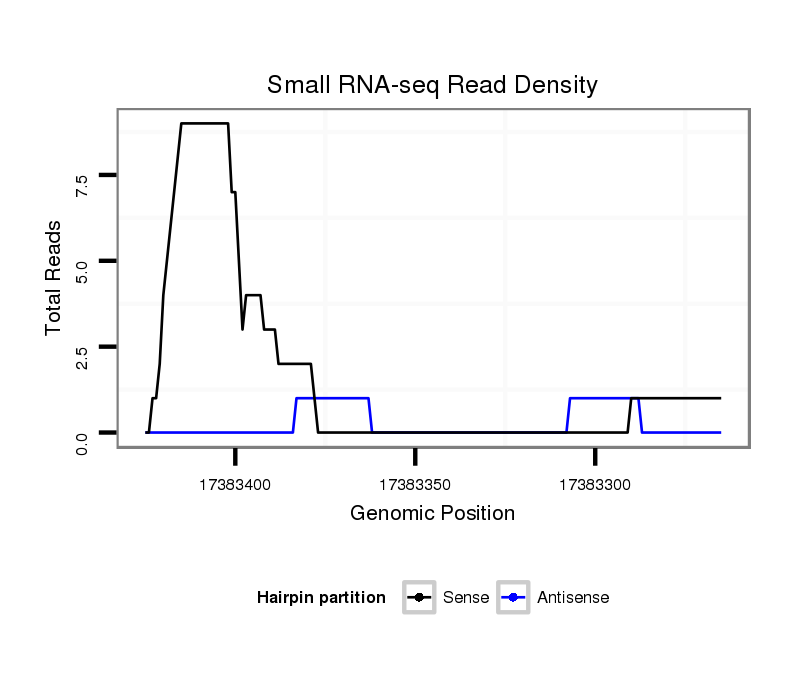
ID:dsi_15164 |
Coordinate:3r:17383315-17383375 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
CDS [3r_17383137_17383314_-]; exon [3r_17382802_17383314_-]; CDS [3r_17383376_17383664_-]; exon [3r_17383376_17383664_-]; intron [3r_17383315_17383375_-]
No Repeatable elements found
| ##################################################-------------------------------------------------------------################################################## CCCTTCGACTATAGCAAACTGGACACCTACGATATCCTAAACAGGGCCAGGTCAGGCGATTCATTCATCCATCACTAGCGTTATATTGAAGATGCCCTCTATGTTTCACAGGTACCATGTGGATCGCAGCGACTTCCTGCAGGCGCTCAAGTACATGAATC **************************************************....((((.((((.......................))))..))))...............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
SRR553485 Chicharo_3 day-old ovaries |
M053 female body |
M025 embryo |
SRR553486 Makindu_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR902008 ovaries |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....TCGACTATAGCAAACTGGACACC...................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....CGACTATAGCAAACTGGAC......................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................GTACCATGTGGTTCGCAGCGA............................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........TATAGCAAACTGGACACCTACGATATCC............................................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......GACTATAGCAAACTGGAC......................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................ACGATATCCTAAACAGGGC.................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................TTCCTGCAGGCGCTCAAGTACATGAAAA | 28 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..CTTCGACTATAGCAAACT............................................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CCTGCAGGCGCTCAAGTACATGAATC | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......ACTATAGCAAACTGGACAC....................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CTATAGCAAACTGGACACCTACGAT................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........ATAGCAAACTGGACAC....................................................................................................................................... | 16 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....CGACTATAGCAAACTGGACACC...................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................GGACACCTACGATATCCTAAACAGGGCC................................................................................................................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................GCAGGCGCTCAGGTA........ | 15 | 1 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................CCTTCACTAGCGGTATAA........................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GGGAAGCTGATATCGTTTGACCTGTGGATGCTATAGGATTTGTCCCGGTCCAGTCCGCTAAGTAAGTAGGTAGTGATCGCAATATAACTTCTACGGGAGATACAAAGTGTCCATGGTACACCTAGCGTCGCTGAAGGACGTCCGCGAGTTCATGTACTTAG
**************************************************....((((.((((.......................))))..))))...............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................TCCCGGTCCAGTCCGCTAAGT.................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CACCTAGCGTCGCTGAAGGA....................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................AAGTGTCCATGATAC.......................................... | 15 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................TCCGTTAAGTAAGTAG............................................................................................ | 16 | 1 | 11 | 0.18 | 2 | 0 | 0 | 0 | 0 | 2 | 0 |
| GGGAAGCTGATATTG.................................................................................................................................................. | 15 | 1 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................CAATATAACTGGTACGG................................................................. | 17 | 2 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................CCGCGATTTCATGGACT... | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................CAGTCCGTTAAGTAA................................................................................................ | 15 | 1 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................AGTGTCCATGATACT......................................... | 15 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
Generated: 04/23/2015 at 05:49 PM