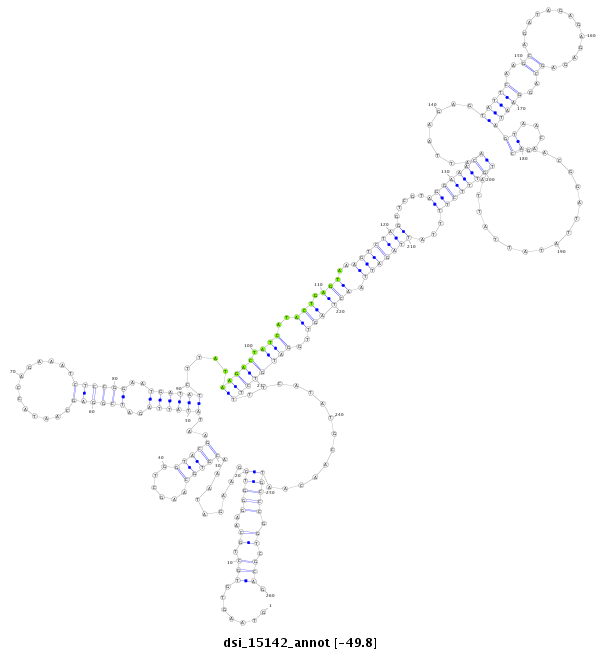
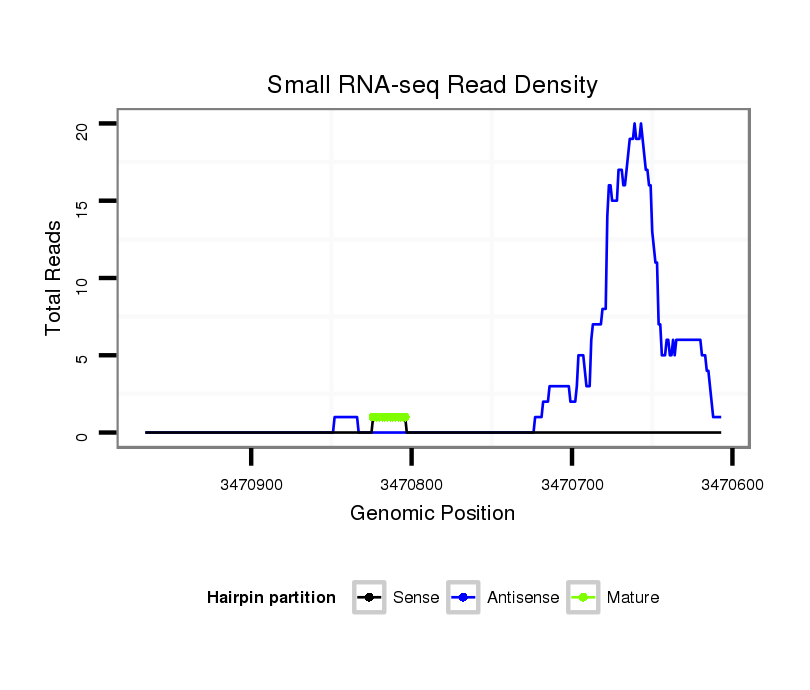
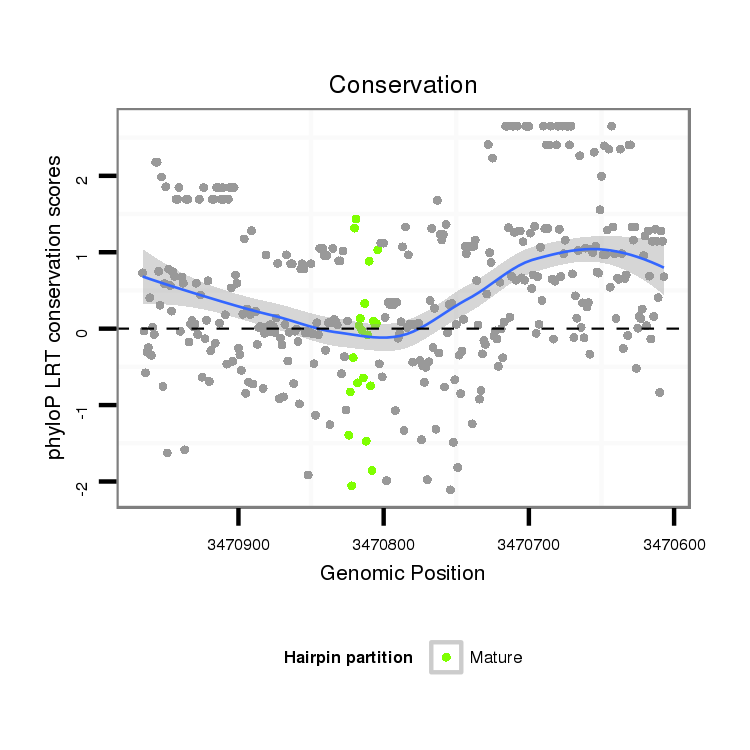
ID:dsi_15142 |
Coordinate:x:3470657-3470916 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -49.6 | -49.5 | -49.5 |
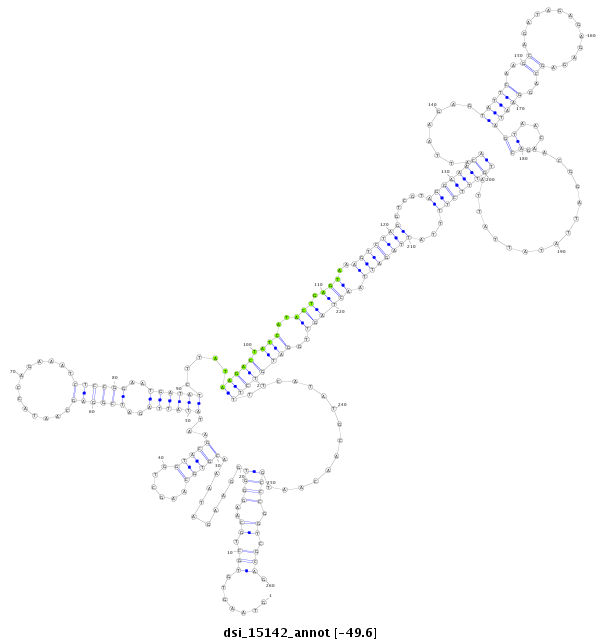 |
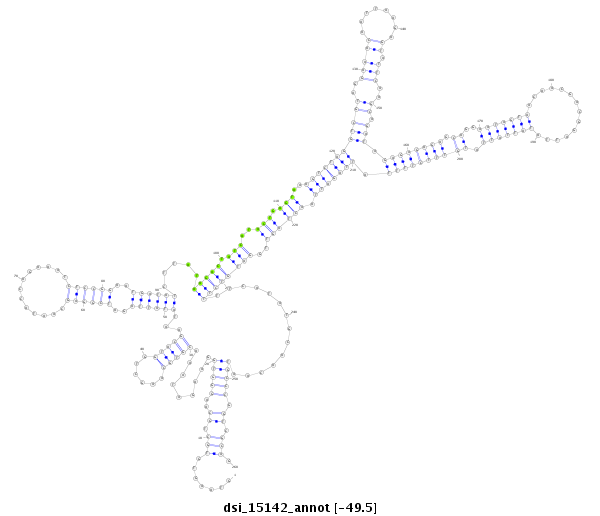 |
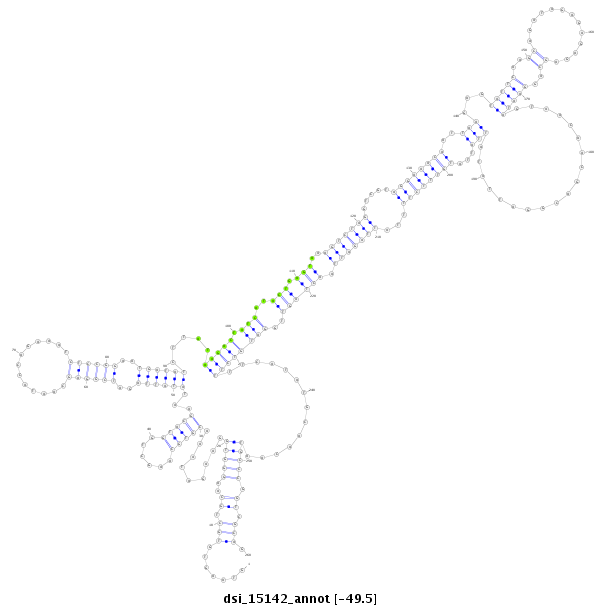 |
Antisense to three_prime_UTR [x_3470530_3470732_+]; Antisense to exon [x_3469540_3470732_+]
No Repeatable elements found
|
ATACCGCTGAAGACGCGTAGCTGGGTATTACCACTGCTCGGTATCAATGGGTAAGTGTGCTGCAAGGGTGGAAGATAAACGTGCAAGCTGGTACGAATATATTAGATCGGAGCAATACCAGAAATCTCCGGAATGATATCTTATAAGACTATCATACTGAGTAAAGTCTAGGTCGTAGGAAACAATTAAGAGTATTCAAGCAGATAGAGAGAGAGCAGGAATAGTAACAACGACGGATTATATTATTATGTTTCTTTTATTAGATTAACTAGTTGGATGTCTTTTCATATGCAACAATGCCCGGTCGCAGCTTCACTTACACTTAGCTTGGAAAAGGACAAAGGCCACTAAGGATTTACA
**************************************************.......(((.((..(((((.........(((((......)))))...((((((..((((((.............))))))..)))))).....(((((.(((..(((.(((..(((((((.....((((((((........(((((..((.............))..)))))((....)).................))))))))...))))))).))))))..))))))))..............))))).)).))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM343915 embryo |
SRR902008 ovaries |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................TGAATGGGTAAGTCTGCTGCAATGG.................................................................................................................................................................................................................................................................................................... | 25 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................ATAAGACTATCATACTGAGTA..................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................TATTAGAGTAACTAGTTGTATG................................................................................. | 22 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................TCTTTGGTTAACTAGTTGGATG................................................................................. | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................GGAAACAATCAAGAGTA...................................................................................................................................................................... | 17 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................................................GAGAGAGCCGGAATCGTA...................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................ATCAGATTAGCTAGTTGG.................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................GGTGCAAGCTGGTAGGAA....................................................................................................................................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................CGAATATATTAGCACGGA......................................................................................................................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................TATTAGTTTATCTAGTTGTATG................................................................................. | 22 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........GACGACGCGTAGCTG................................................................................................................................................................................................................................................................................................................................................. | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................TATATTAGATTAGCTTGTTGG.................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......CTAAGACGCGTAGCTGGT............................................................................................................................................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .......................................................................................................................AGAAGTCTCCTGAATGA................................................................................................................................................................................................................................ | 17 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................................................................................CACATGCTACAATGGCCGG........................................................ | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................ACATGCTACAATGGCCGGT....................................................... | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................................................................................................................CAGATTAGCTAGTTGG.................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............GCGGAGCTGGGTTTTACCG....................................................................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................GGTGCTGGAAGGGTGGA................................................................................................................................................................................................................................................................................................ | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................ATGCTACAATGTCCGGTA...................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------############################################################################################################################## TATGGCGACTTCTGCGCATCGACCCATAATGGTGACGAGCCATAGTTACCCATTCACACGACGTTCCCACCTTCTATTTGCACGTTCGACCATGCTTATATAATCTAGCCTCGTTATGGTCTTTAGAGGCCTTACTATAGAATATTCTGATAGTATGACTCATTTCAGATCCAGCATCCTTTGTTAATTCTCATAAGTTCGTCTATCTCTCTCTCGTCCTTATCATTGTTGCTGCCTAATATAATAATACAAAGAAAATAATCTAATTGATCAACCTACAGAAAAGTATACGTTGTTACGGGCCAGCGTCGAAGTGAATGTGAATCGAACCTTTTCCTGTTTCCGGTGATTCCTAAATGT **************************************************.......(((.((..(((((.........(((((......)))))...((((((..((((((.............))))))..)))))).....(((((.(((..(((.(((..(((((((.....((((((((........(((((..((.............))..)))))((....)).................))))))))...))))))).))))))..))))))))..............))))).)).))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M053 female body |
M024 male body |
M025 embryo |
GSM343915 embryo |
O001 Testis |
SRR553485 Chicharo_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................................................................................................TACGTTGTTACGGGCCAGCGTCGAAGTG............................................ | 28 | 0 | 1 | 3.00 | 3 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................CAGAAAAGTATACGTTGTTA.............................................................. | 20 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................TTACGGGCCAGCGTCGAAGTGAATG........................................ | 25 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................CGAAGTGAATGTGAATCGAAC.............................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................CAGGCCAGCGTCGAAGTGAATG........................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................CCAGCGTCGAAGTGAATG........................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................CCAGCGTCGAAGTGAATGTG...................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................GCGTCGAAGTGAATGTGAA.................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................TCGAAGTGAATGTGAATCGAACCTTT.......................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................ACAAAGAAAATAATCTAATTGATCAA...................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................GCCAGCGTCGAAGTGAATGTGAATCG................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................TACGTTGTTACGGGCCAGCGTCGA................................................ | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................AACCTTTTCCTGTTTCCGGTGATTCC....... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................CGGGCCAGCGTCGAAGTGAATG........................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................GTCTTTAGAGGCCTT................................................................................................................................................................................................................................... | 15 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................CGTTGTTACGGGCCAGCGTCG................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................TACGTTGTTACGGGCCAGCG.................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................ACGTTGTTACGGGCCAGCGTC.................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................GGCAGCGTCGAAGTGAATGTGA..................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................AGAAAATAATCTAATTGATCAAC..................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................TCCTGTTTCCGGTGATTCCTAAATGT | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................GGCCAGCGTCGAAGTGAATGTG...................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................AGAAAAGTATACGTTGTTACGGGCCAG...................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................TTTTCCTGTTTCCGGTGATTCCT...... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................TCAACCTACAGAAAAGTATACGTTGTTA.............................................................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................TCAACCTACAGAAAAGTATAC..................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................ATCAACCTACAGAAAAGTATA...................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................TACGTTGTTACGGGCCAGCGTCGAAG.............................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................TCGAACCTTTTCCTGTTTCCGGTGATTC........ | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................ATAATACAAAGAAAATAATCTA............................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................CGGGCCAGCGTCGAAGTGAA.......................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................CAGAAAAGTATACGTTGTTACGGG.......................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................CCTTTTCCTGTTTCCGGTGAT.......... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................CGTCGAAGTGAATGTGAATCG................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................ACGTTGTTACGGGCCAGCGTCGAAGTGA........................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................GTATACGTTGTTACGGGCCAG...................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................CGAACCTTTTCCTGTTTCCGGT............. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................TTCTGTTTGTACGTTCGACTAT........................................................................................................................................................................................................................................................................... | 22 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................................CTGCCCAATAAAATAATA............................................................................................................... | 18 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................ATGTAGAGGCCTTACT................................................................................................................................................................................................................................ | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................................................................................ATTGTTGCTTCCTAATACT..................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | x:3470607-3470966 - | dsi_15142 | ATACCGCTGAAGACGCGTAGCTGGGTATTACCACTGCTCGGTATCAATGGGTAAGTGTGCTGCAAGGG-TGGAAGATAAACGTGCA-------AGCTGGTAC-------------GAATATA-------------------------------------------------------------------------------------------------------------------------------------------------------TTAGATCGGAGCAATACC-------------AGA-------------------------------------------AATCTCCGGAATGATAT-CTTATA--AGACTAT-----------------CATAC-----------------TGAGTAAAGTCTAGGTCGT-------AGGAAACAATTAAGAGTATTC-A--AGCAGA------------TAGAGAGAGAGCAGGAATAGTAACAA---CG--------------------------------------------------------------ACGGA--TTATATTATTATGTTTCT-TTTATTAGATTAAC--T-------AGT----TGGATGTCTTTTCATATG-CAACAATG-C------CCGG----T-----------------------------------------------CGCAGCTTCACTTACACTTAGCTTGGAAAAGGACAA--AGGCCAC-------------------------T--AAGGATTTACA |
| droSec2 | scaffold_4:2934457-2934816 + | ATACCGCTGAAGACGCGTAGCTGGGTATTACCACTGCTCGGTATCAATGGGTAAGTGTGCTGCAAGGG-TGGAAGATAAACGTGCA-------AGCTGGTAC-------------GAATATA-------------------------------------------------------------------------------------------------------------------------------------------------------TTAGATCGGAGCAATACC-------------AGA-------------------------------------------AATCTCCGGAATGATAT-CTTGTA--AGACTAT-----------------CATAC-----------------TGAGTAAAGTCTAGGTCGT-------AGGAAACAATTAAGAGTATTC-A--AGCAGA------------TAGAGAGAGAGCAAGAATAGTAACAA---CG--------------------------------------------------------------ACGGA--TTATATTATTAAGTTTCT-TTTATTAGATTAAG--T-------AGT----TGGATGTCTTTTCATATG-CAACAATG-C------CCGG----T-----------------------------------------------CGCAGCTTCACTTACACTTAGCTTGGAAATGGACAA--AGGCCAC-------------------------T--AAGGATTTACA | |
| dm3 | chrX:3772732-3773098 - | ATACCGCTGAAGACGCGTAGCTGGGTATTACCACTGCTCGGGATCAATGGGTAAGTGTGCTGCAAGGG-TGGAAGATATACGTGCA-------AGCTGGTAC-------------TAATATA-------------------------------------------------------------------------------------------------------------------------------------------------------TTAGATCGGAGCAATACC-------------AGA-------------------------------------------AATCTCCGGAATGATAT-CGTATA--AGACTAT-----------------CATAC-----------------TGAGTAAAATCCAGGTCATAGGTGAAAAGAAACAATTAAGAGTATTA-A--AGCAGA------------TAGAGAGACAACAAGAATAGTAACAA---CG--------------------------------------------------------------ACGGA--TTATATTATTAATTTTCT-TTTATTAGATTAAC--T-------AGT----TGGATGTCTTTTCATATG-CAACAATG-C------CCGG----T-----------------------------------------------CGCAGCTTCACTTACACTTAGCTTGGAAATGGACAA--AGGCCAC-------------------------T--AAGGATTTACA | |
| droEre2 | scaffold_4690:1156701-1157038 - | ATACCGCTGAAGACGCGAAGCTGGGTATTACCACTGCTCGGGATCACTGGGTAAGTGTGCTGCAAGGG-TGGAAAATATACGTGCA-------AGCTGGTAC-------------TAATATA-------------------------------------------------------------------------------------------------------------------------------------------------------TTAGATCAGAGCAGTACC-------------AGA-------------------------------------------AATCTCCGGAATGATAT-TTTCTA--AGAGTAT-----------------TT-TC---------------------------------------TGGAGAGACACATTTAAAAGA-----G--AGT--A------------TAGAGAGACATCAAGAATAGTAACAA---CG--------------------------------------------------------------ACGGA--TTATAGTATTAATTTTCTTTTTATTAGATTAAC--T-------AGT----TGGATGTCTTTTCATATG-CAACAATG-C------CCGG----T-----------------------------------------------CGCAGCTTCACTTACACATAGCTTGGAAA-GGACAG--AGGCCAC-------------------------T--AAGGATTCACA | |
| droYak3 | X:4642029-4642391 + | ATACCGCTAAAGACGCGAAGCTGGGTATTACCACTGCTCGGGATCAATGGGTAAGTGTGCTGCAGGGG-TGGAAGATATACGTGCA-------AGCTGGTAC-------------TAATATA-------------------------------------------------------------------------------------------------------------------------------------------------------TTAGAACAGAGCATTACC-------------AGA-------------------------------------------AATCTCCGGAATGATAT-TTTATG--AGACTAT-----------------GA-AC-----------------TACGTAAAATCGAGGTCATAGATGGAGAGACACATTCAAGGATATTT-A--AGCAGA------------TAGACAGACAGCAGGAATAGTAACAA--------------------------------------------------------------------CGAA--TTATATTTTTACTTTTCT-TTTATTAGATTAAC--T-------AGT----TGGATGTCTTTTCATATG-CAACAGTG-C------CCGA----T-----------------------------------------------CGCAGCTTCACTTACACTTAGCTTGGAAATGGACAA--AGGTCAT-------------------------T--AAGAATTCACA | |
| droEug1 | scf7180000409553:23153-23594 - | ATACCGCTAAAGACTCGAAGTTGGGTATTACCACTACTCGGAATCAATGGGTAAGTGTGCTGCAAAGG-ATGGAAATATAGATAGG-------AACTCCTA-----------TAT--ACATA-----------------------------GAACCA-------------------------AAAC--------------------------TCCTATTATTGGATA----CCTCTATTCTTCTCTGTACTACTTAAATCCTTCCTAGCATTTATACATTCGAGCACCATG-------------CAA-------------------------------------------AATCACCAGATTTATAT-TTTAATACGGACTTC-----------------AATAC-----AGAGTATTACAGATAGCACAATTT-------AGATAGAAAAATACATCTAAGAACAAA--A--AGCATACATAGGATAG-------------A--ATATAGTAATAACAC------------------------------------------------------GCA-------ACGAG-TTTATATTAATTGTTTTCT-TTTATTAGATTAAC--T-------AGA----TGGATGTCTTTTCATATG-CAACAACG-C------CCGG----T-----------------------------------------------CGGAGCTTCACTTACACTTAGCTTGGGAATGGACAG--AGGCCAC-------------------------T--AAGGATTTACA | |
| droBia1 | scf7180000302421:2544845-2545207 - | ATGCCGCTGAAGACCCGCAGCTGGGTGTTCCCTCCACTGGGGATCAGTGGGTACGTGTGCTGCAAGGGATGGAAGATGTAA--GG-------------GTGCTCCC-----AAGT--GC-----------------------------------------------------------------------------------------------------------T----T----------------------------------------------------------TGCCACTCCCCCCCGGGAGAGCAGCCGGCTCTCCACTCTTCTATGATATCTTGATACTATAATCTCCGGAATGATGT-TTTGGC--AGAG-------------------------------------------------AGTTC-------AGATAGAGA--------------------A--AGCGTA------------CAGAGAG----T--GCGGAGTAAGAACAGCG------------------------------------------------------A-------GCGGG--TTATATCAGTGGTTTTCT-TTTATTAGATTAAC--T-------AGA----TGGATGTCTTCTCATATG-CAACAGCG-C------CCGG----T-----------------------------------------------CGGGGCTTCACTTACACCTAGCTCGGGAATGGACAG--GGGCCCC-------------------------TGGGGGGACTCACA | |
| droTak1 | scf7180000415248:568619-568939 + | ATGCCGCTGAAGACCCGGAGCTGGGTATTGCCACTACTCGGGATCAGCGGGTAAGTGCGCTGCAAGGA-AGGTATAT-------------------------TCCCTATATAAGT--A-----------------------------------------------------------------------------------------------------------TT----C----------------------------------------------------------CCCTACTATTCTCATGG-------------------------------------------AATCTCCTGAATGATCTTCCTATA-GAGACTAA-----------------CACA------------A------GATCACAGTTT-------AGAAAGAAAGA----------------------------------------------------------------ACAGCGAAAGACAGACAAA------------------------------------CAAGCA-------GCGGG--CTATAATAATAGTTTTCT-TTTATTAGATTAAC--T-------AGA----TGGATGTCTTTTCATATG-CAACAACG-C------CCGG----------------------------------------------------CGGGGCTTCACTTACACTTAGCTTGGAAATGGACAG--AGGCCAC-------------------------T--AAGGATTCACA | |
| droEle1 | scf7180000491018:207980-208086 + | ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATATCAATAGTTTTCA-TTTATTAGATTAAC--C-------AGA----TGGATGTCTGGTCATATG-CAACAACG------------------------------------------------------------------------CACTTACACTTAGCT-GGGAATGGACAG--TGGCCAC-------------------------T--AAGGATTCACA | |
| droRho1 | scf7180000779334:961693-962145 + | ATACCGCTGAAGACCCGAAGCTGGGTATTACCACTACTCGGGACCAGGGGATAGGTGTTCTGGAAGA-----------TACATGCAGTTTCAGAGCTGGAAC-------------TCCTATTTAAATTACTGTGAGAACATTTCGTTGAGAAAACCACTTCAATTGGGGATGTTGGAAATACAAACTTATTGAGGACTACTTCAGAATTGAATCTAATTATTGGACAATGGTCTCTATTGCT------------------------------------------------------------CA--------------------------------------------------------------T-TGAAATAGGGACTAT-----------------CAGACTATACAGAGTGGTACACAAAGCACAGTTC-------AGATAGAAAGAAATATTCTAGAGATGTG-AACAGCAGA------------CAGACAAAATATATATATAGTAACAACAC------------------------------------------------------GCA-------GCGGG--TTATATCAATAGTTTTCT-TTTATTAGATTAAC--T-------AGA----TGGATGTCTTGACATATG-CAACAAC------------------------------------------------------------------------TCACTTACACTTAGCTTGGGAATGGACAA--GAGCCAC-------------------------T--AAGGATTCACA | |
| droFic1 | scf7180000453901:696419-696782 + | ATTCCGCTGAAGACCCGCAGTTGGGTGTTGCCAATACTCGGGATCAGGGGATAGGTGTTCTGAAAGAA-A----------CAG------TCAGAGTCGGAACTCCC-A----CATGAAGTTG-------------------------------------------------------------------------------------------------------------------------------------------------------TTAAGTTCGAGTGATACCCAAACTAGCATC-TGG-------------------------------------------AATCCCTCAAAGG-------------AGACCAT-----------------CAAGCGTTACA-----GCACAGCTAGCAAAGTTC-------AGATAGAAAGACACATTTAGGAGCGAC--A--AG---------------------AAGGAGC--ACGTAGTAAGAACGC------------------------------------------------------ACA-------TGCAG--TTATATCGGTAGTTTTCT-TTTATTTGATTAAC--C-------AGA----CGGATGTCTTGTCATTTG-CAACAACG-C------CCGG----T-----------------------------------------------CAGAGC-TCACTTACACTTAGTTTGGGAATGGACAG--AGGCCAC-------------------------T--AAGGATTCACG | |
| droKik1 | scf7180000302517:57701-57879 + | GATTACAC-AAGCC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTAAGCAG-----------------------------------------------------GAG-----------CAG-AAAATATAGAG--G------------------------------------AGAGCC-------ATAAA--TTGTATCAATAGTTTTCT-TTTATTATTTTAAC--T-------AAAT---TGGATGTCTTGTCATATG-CAACAACG-CCGCC--CCAG----T-----------------------------------------------TGGGGCTTCACTTACACTTAGCCTGGTTATGGACAG--AAGCCAC-------------------------T--AAGGATTCACA | |
| droAna3 | scaffold_13117:711557-711844 - | ATGCCGCTGAAGACCCGCAGCTGGGTGTATCCGCCCGAGGCTACTTCGGGGTAGGTGTTCTAGAAGGA--GGAGGGTGTTC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TG--GGAGC-TCAGGCAGGAGAGACAAACACA------------GGAC---------------------------------A--------------CAC--GGGATACGGGCGACAGGA--------------------------------------------------CACACAAACATCGCAGAGCGGCAGAGCGGAAGAGCG-------GAAGA--GTATATCGGTAGGTTTCT-TTTATTGG-TTAACATT-------TGG----TGGAGGTCTGGTCAGGTG-CAACAAGG-G------CCTG----G--AGCG-----------------------------------------CGGGGC------------TGGCCA-GGATGGGACAG--AGACCAC-------------------------T--AGGAC--TACG | |
| droBip1 | scf7180000396412:630580-630862 - | ATGCCGCTGAAGACACGCAGCTGGGTGTACCCGCCCGAGGCTATTACAGGATAGGTGTCCTAGAAGA--TGGGGGATC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCAGACGGG-------------------------GGAC---------------------------------A--------------CAG--GGGACACAGGGGACAGGA--------------------------------------------------CACACAAACATAGCAGAGCAGCAG--------AGCA-------GCAGG-TGTATATCGGTAGGTTTCT-TTTATTAGGTTAACATT--TGGA-TTG----TGGATGCCTGGTCAGGTG-CAACAACG-GGGCATCCCTG----A-----------GCAGCCAAGCT-------------------------CGGGGCTGGG-----GC-TGGACAGGGTGGGGACAG--GGACCAC-------------------------T--AGGAC--TACG | |
| dp5 | XL_group1e:6918091-6918263 - | ACAGAGGA-AAGGCAGGAGGCGA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGGCTTT---CTAACAGTTTTCTTTTTATTAGATTAAC--T-------AAA----TGAATGTCTTCTCATATG-CAACAACA-G------CCCG----A-----------------------------------------------CAGAGCTTCACTTACACTTAGATTGGAAATGGACAC--AGGCTATATGGATCGCATCGTAATCGTAATCGT--AGGGATTGACA | |
| droPer2 | scaffold_25:254696-254830 - | ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCTT-TTTATTAGATTAAC--T-------AAA----TGAATGTCTTCTCATATG-CAACAACA-G------CCCG----A-----------------------------------------------CAGAGCTTCACTTACACTTAGATTGGAAATGGACAC--AGTCTATATGGATCGCATCGTAATCGTAATCGT--AGGGATTGACA | |
| droWil2 | scf2_1100000004909:11866156-11866313 - | AT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATATTTTA-TTCAGTAGATTAAC--T-------AGAA---AGAATGTCTTGTCATATTATAACAACG-GTGT---CCGG----A-----------GCATCCGAACATGTTACTTTTGTGTGT---------CAC-ACTT----CATACTTACATTGTAAAAGGGTGAACAGGGTATGGGGGT---A---------TGGGGGT--GGGGATGAACA | |
| droVir3 | scaffold_13042:4658925-4659140 - | AGAGAGAGA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAGAGCGAG-----------GGG-AAGATAAAGAC----AAACACAC------------------------------GCAAACAACTAT-GA--TTACAATTTTTGATTTATTTGTATAAGATTAGT--TTAGTT--AAAA---CGTATGTCTTGGCATATT-CAACAAGG-C------CCGTCCCGTCGATCTCTGGGTCCG-----------------ATCG-TTGACCCGGGCAGGGTT----TTACACTTACATTGTAAATGGACAC--AAAGTACAT-----------------------A-TATGAATTATCA | |
| droMoj3 | scaffold_6328:3923274-3923454 - | TCAAGAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A------------AAGAGAGATA--------------------------AAGA--GACGAACAC--------------------------------ACA----GG-TATGA--TTATAATTTCGTATTTAT-TTAATTAGATTAGT--TTAATT-TAAAGCGTACTATGTCTTTTCATATT-CAACAAACGC------TCG-----ACGACCCCTAAAACCG----------------------T---------C-GAGCTTC----ACACTTACATTGT-AAGGGACAC--AAA---T-------------------------T--AAAAATATATA | |
| droGri2 | scaffold_15081:2841243-2841308 + | ATGCCGCTAAATACACGCACCTGGGTGCTGCCCATCTTGGGCACAACGGGGTAGGTGCGCTGCAAG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/19/2015 at 05:53 AM