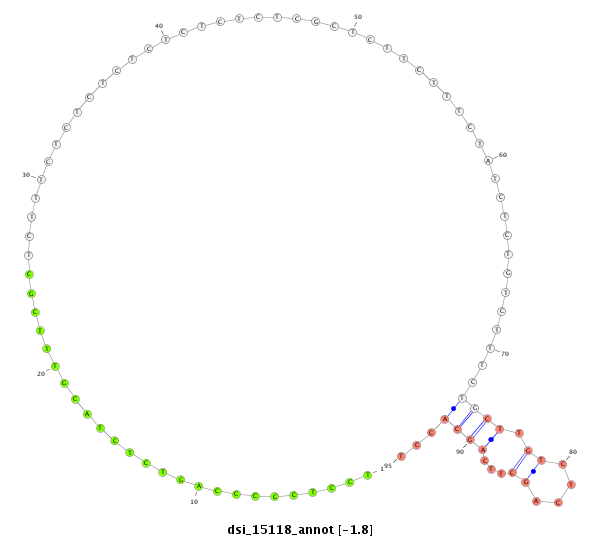
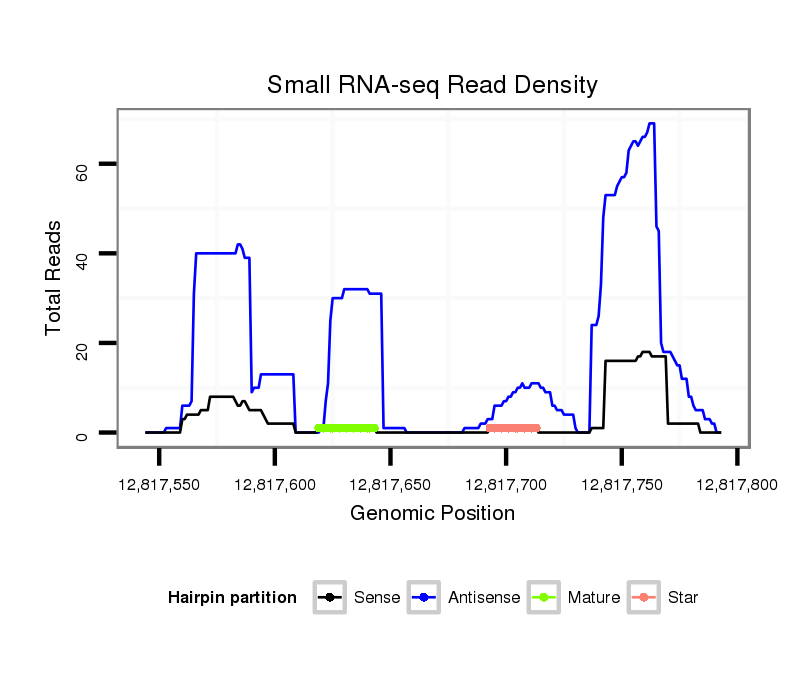
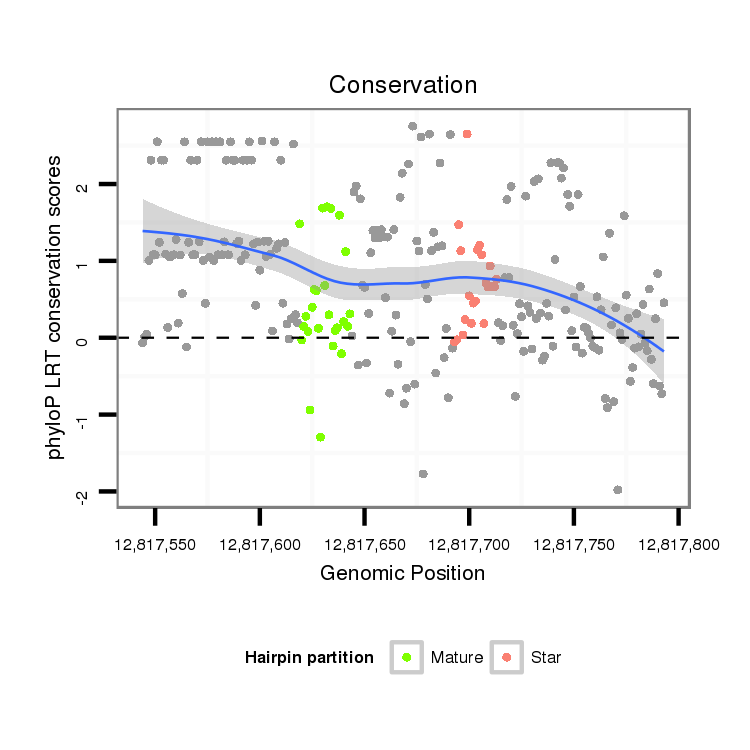
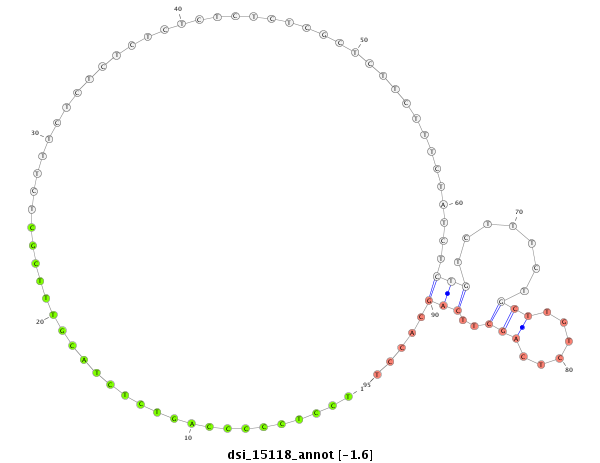
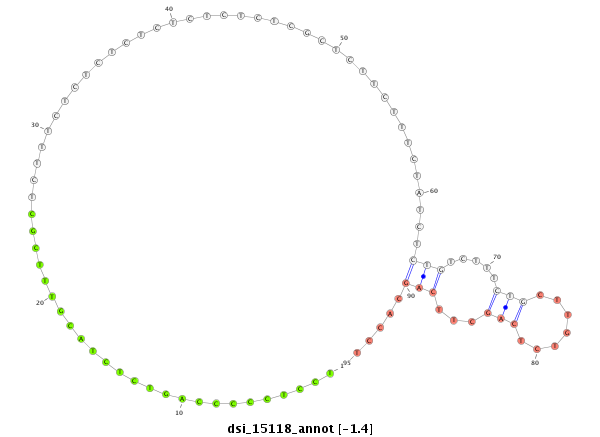
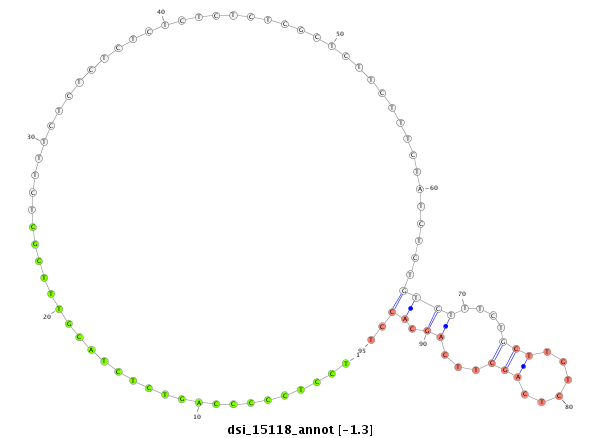
| .....................TTGGCCTACGTCAGTGACACCACGT............................................................................................................................................................................................................ |
25 |
0 |
1 |
24.00 |
24 |
18 |
5 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................AGACTTAGTAAGCTAAAGGGTGTCAGGT............................. |
28 |
0 |
1 |
20.00 |
20 |
15 |
5 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................GGGGTCAGAGATGCAAAGCGAGA................................................................................................................................................... |
23 |
0 |
1 |
14.00 |
14 |
13 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................TAGTAAGCTAAAGGGTGTCAGGTGT........................... |
25 |
0 |
1 |
13.00 |
13 |
10 |
2 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................TGGCCTACGTCAGTGACACCACGT............................................................................................................................................................................................................ |
24 |
0 |
1 |
9.00 |
9 |
7 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................CGTGGATAAAGTAACGATGGGTAAGT............................................................ |
26 |
2 |
1 |
8.00 |
8 |
6 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................TGGCCACTCAGAACAGCAT......................................................................................................................................................................................... |
19 |
0 |
1 |
6.00 |
6 |
6 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................AGGGGGTCAGAGATGCAAAGCGAGA................................................................................................................................................... |
25 |
0 |
1 |
6.00 |
6 |
5 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................TTAGTAAGCTAAAGGGTGTCAGGTGT........................... |
26 |
0 |
1 |
6.00 |
6 |
5 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................GGGTCAGAGATGCAAAGCGAGA................................................................................................................................................... |
22 |
0 |
1 |
5.00 |
5 |
3 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................................AGGGTGTCAGGTGTTGAACACGTAAT............... |
26 |
0 |
1 |
4.00 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................GGGGGTCAGAGATGCAAAGCGAGA................................................................................................................................................... |
24 |
0 |
1 |
4.00 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................................................AGATAGACTTAGTAAGCTAAAGGGTGT.................................. |
27 |
1 |
1 |
4.00 |
4 |
2 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................................................................AGTAAGCTAAAGGGTGTCAGGTGT........................... |
24 |
0 |
1 |
4.00 |
4 |
1 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................CACTCAGAACAGCAT......................................................................................................................................................................................... |
15 |
0 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................AGAACGAGACAGAAAGACGAACAGAGT............................................................................................ |
27 |
2 |
1 |
3.00 |
3 |
2 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................................................AGATAGACTTAGTAAGCTAAAGGGT.................................... |
25 |
1 |
1 |
3.00 |
3 |
2 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................................................................AGTAAGCTAAAGTGTGTCAGGTGT........................... |
24 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................CCACGTTGGCCACTCAGAACAGCAT......................................................................................................................................................................................... |
25 |
0 |
1 |
2.00 |
2 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................ACCACGATGGCCACTC................................................................................................................................................................................................... |
16 |
1 |
2 |
2.00 |
4 |
0 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................CTTAGTAAGCTAAAGGGTGTCAGGT............................. |
25 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................ACAGAGTCGAAGTCGTGGATAAAGT.......................................................................... |
25 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................AAGTTTTGGCCTACGTCAGTGACACCACGT............................................................................................................................................................................................................ |
30 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................................................................................................................GGTGTTGAACACGTAATATTATATTATGT... |
29 |
0 |
1 |
2.00 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................AGACTTAGTAAGCTAAAGGGTGTCAGGTGT........................... |
30 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................AAGTTTTGGCCTACGTCAGTGACACCA............................................................................................................................................................................................................... |
27 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................AGACGAACAGAGTCGAAGTCGTGGAT............................................................................... |
26 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................AGAGAACGAGACAGAAAGACGAAC................................................................................................. |
24 |
3 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................TCGTGGATAAAGTAACGATGGGTA............................................................... |
24 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................................................................................TAATAGATAGACTTAGTAAGCTAAAGGG..................................... |
28 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................GGGTGTCAGAGATGCAAAGCGAGA................................................................................................................................................... |
24 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................ATGGCCACTCAGAACAGCAT......................................................................................................................................................................................... |
20 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................................................AGTAATAGATAGACTTAGTAAGCTAA......................................... |
26 |
2 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................................GACAGAAAGACGAACAGAGTCGAAGT...................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................GAAGTCGTGGATAAAGTAACGATGGGT................................................................ |
27 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................AGAGAGAACGAGACAGAAAGACGAAC................................................................................................. |
26 |
3 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................................................................AGTAAGCTAAAGGGTGTCAGGTGTT.......................... |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................AGACTTAGTAAGCTAAAGGGTGT.................................. |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................AAGTTTTGGCCTACGTCAGTGACACC................................................................................................................................................................................................................ |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................AGAGATGCAAAGCGAGA................................................................................................................................................... |
17 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................GGGGGTCAGAGATGCAAAGCGTGT................................................................................................................................................... |
24 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................CGTGGATAAAGTAACGATGGGTAATT............................................................ |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................TCGAAGTCGTGGATAA............................................................................. |
16 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................................................................CTAAAGGGTGTCAGGTGTTGAACACGT.................. |
27 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................................................................................GCTAAAGGGTGTCAGGTGTTGAAC...................... |
24 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................GAACGAGACAGAAAGACGAACAGAGT............................................................................................ |
26 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................................AGGGTGTCAGGTGTTGAACACGT.................. |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................................................................TAGATAGACTTAGTAAGCTAA......................................... |
21 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................TTTAGTAAGCTAAAGGGTGTCAGGTGT........................... |
27 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................AGAGATGCAAAGCGAGAAAGAGAGAGA......................................................................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................................GGATAAAGTAACGATGGGT................................................................ |
19 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................................................................................TCAGGTGTTGAACACGTAATAT............. |
22 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................AGTCGTGGATAAAGTAACGATGGGT................................................................ |
25 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................CGTGGATAAAGTAACGATGGGTAAG............................................................. |
25 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................TAGTAAGCTAAAGGGTGTCAGGT............................. |
23 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................................AAGGGTGTCAGGTGTTGAACA..................... |
21 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................GGTGTCAGGTGTTGAACACGTAATATT............ |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................AGACTTAGTAAGCTAAAGGG..................................... |
20 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................CGAACAGAGTCGAAGTCGTGGATAAAGT.......................................................................... |
28 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................ACAGAGTCGAAGTCGTGGATAAAGTAA........................................................................ |
27 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................................................................................................................CAGGTGTTGAACACGTAATATTATATTAT..... |
29 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................AGGGTGTCAGAGATGCAAAGCGAGA................................................................................................................................................... |
25 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................................................................TAAAGGGTGTCAGGTGTTGAACACGT.................. |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................................AGGGTGTCAGGTGTTGAACACGTAAC............... |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................................................................................GCTAAAGGGTGTCAGGTGTTGAACAC.................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................................................AGATAGACTTAGTAAGCTAAAGGGTTT.................................. |
27 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................................GTCAGGTGTTGAACACGTAATATTATAT........ |
28 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................................................AGATAGACTTAGTAAGGTAAAGGGTGT.................................. |
27 |
2 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................GGAGGGGGTCAGAGATGCAAA......................................................................................................................................................... |
21 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................................................................AAGATAGACTTAGTAAGCTAAAGGTTGT.................................. |
28 |
3 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................AGATTTAGTAAGCTAAAGGGTGT.................................. |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................GACACCACGTTGGCCACTCAGAACAGCAT......................................................................................................................................................................................... |
29 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................................................AGTCGAAGTCGTGGATAAAGTAACGA..................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................AGCACGAGACAGAAAGACGAACAGAGT............................................................................................ |
27 |
3 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................................................................................................................AGGTGTTGAACACGTAATATTATAT........ |
25 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................TGGGGTCAGAGATGCAAAGCGAGA................................................................................................................................................... |
24 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................................AGGGTGTCAGGTGTTGAACACGTAAGAT............. |
28 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................TAGTAAGCTAAAGGGTGTCAGGTGTT.......................... |
26 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................TTTGGCCTACGTCAGTGACACCACGT............................................................................................................................................................................................................ |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................TTAGTAAGCTAAAGGGTGTCAGGTG............................ |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................................GGGTGTCAGGTGTTGAACACGTAATAT............. |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................GAGGGTCAGAGATGCAAAGCGAGA................................................................................................................................................... |
24 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................GGCCACTCAGAACAGCAT......................................................................................................................................................................................... |
18 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........CGCGGCGAAGTTTTGGCCTACGTCAGT...................................................................................................................................................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................CACGAGGGCCACTCAGAAGA............................................................................................................................................................................................. |
20 |
3 |
6 |
0.33 |
2 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
| .......................................ACCACGATGGCCACTCCTAAC.............................................................................................................................................................................................. |
21 |
3 |
4 |
0.25 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................ACCGCGTCGGCCACTCAG................................................................................................................................................................................................. |
18 |
2 |
4 |
0.25 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................................AGGGTGTCAGGTGTCG......................... |
16 |
1 |
5 |
0.20 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................AGCGAGAAAGAGAGAGAGCGA..................................................................................................................................... |
21 |
1 |
7 |
0.14 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| .......................................ACCACGATGGCCACT.................................................................................................................................................................................................... |
15 |
1 |
9 |
0.11 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................................AGAGAGAGAGAGAGAG................................................................................................................................ |
16 |
0 |
20 |
0.10 |
2 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
| ........TCGGGGCGAAGTCTTG.................................................................................................................................................................................................................................. |
16 |
2 |
20 |
0.10 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................................................AGACTCGATGTCGTGG................................................................................. |
16 |
2 |
20 |
0.10 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................ACCACGATGGCCACTCC.................................................................................................................................................................................................. |
17 |
2 |
13 |
0.08 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................................AGAAAAGATGGGAAATAGGT......................................................... |
20 |
3 |
13 |
0.08 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| .........................................................................................................................................................................................................................AGGTGGTGAACACGT.................. |
15 |
1 |
14 |
0.07 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................TGGCCACTCAGAAGA............................................................................................................................................................................................. |
15 |
1 |
15 |
0.07 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................................................................................CAGAGTCGCAGTCCTG.................................................................................. |
16 |
2 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................AGCGAGAAAGAGAGAGAGTT...................................................................................................................................... |
20 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................................................................................TAATAGGAAGGCTTGGTAA.............................................. |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
| ............................................................................................................................................................................AAGTAACGATGGGTAAGTA........................................................... |
19 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................................................................................AATAGATAGACTTAG................................................. |
15 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |