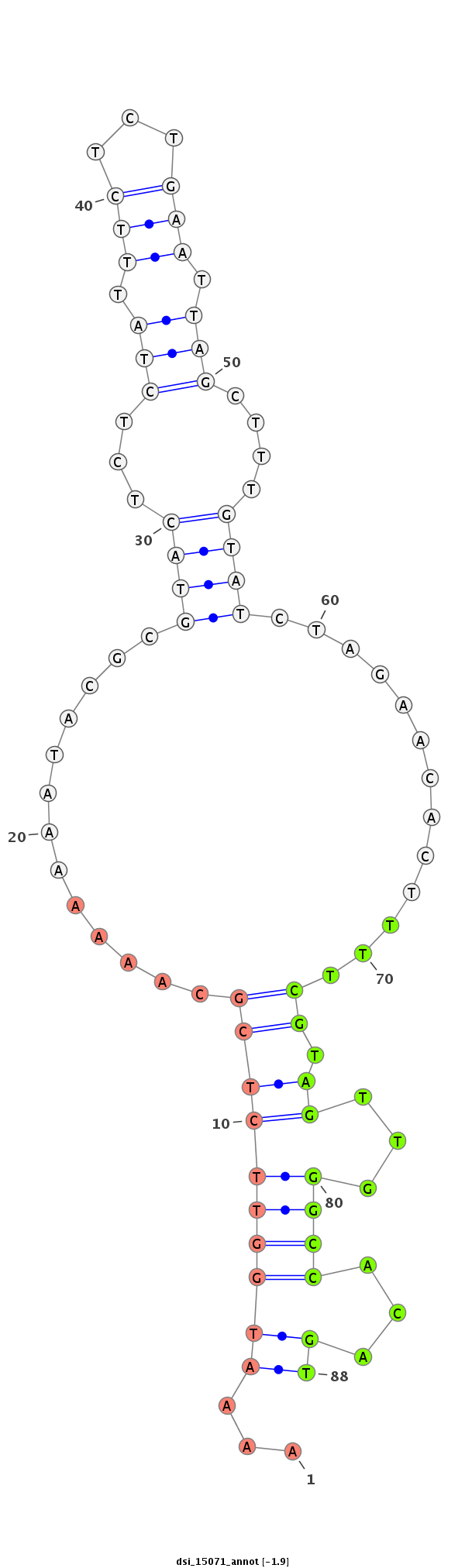
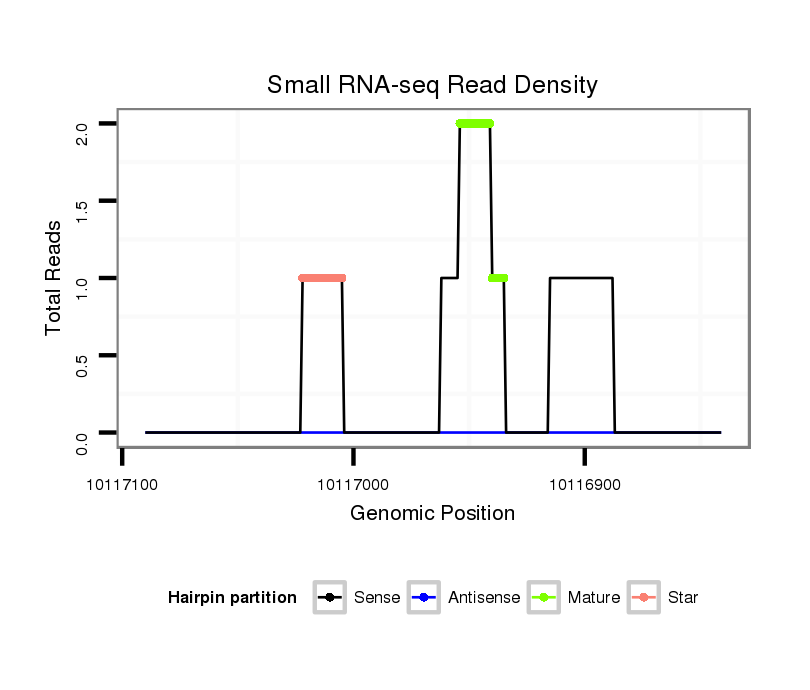
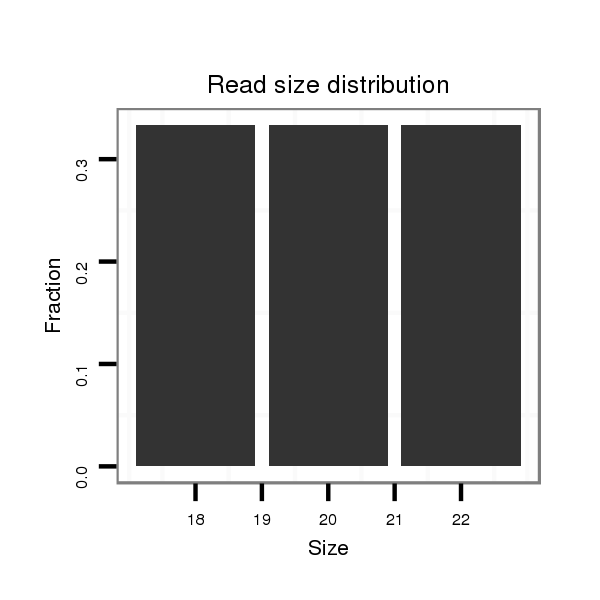
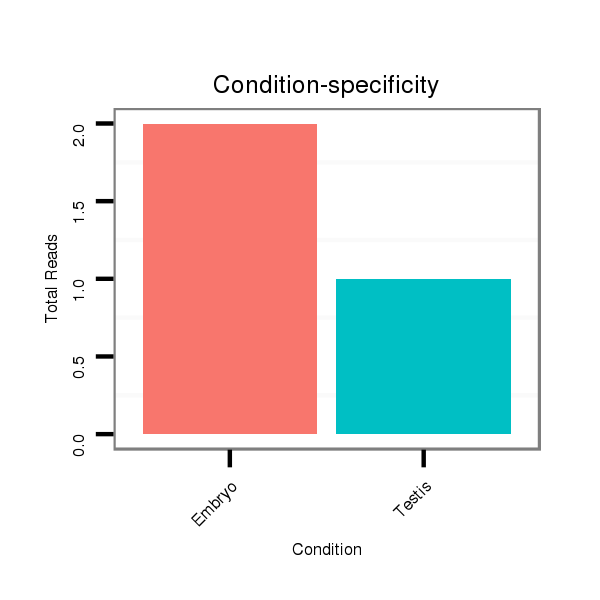
ID:dsi_15071 |
Coordinate:3r:10116891-10117040 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -9.7 | -9.4 | -9.0 | -8.8 |
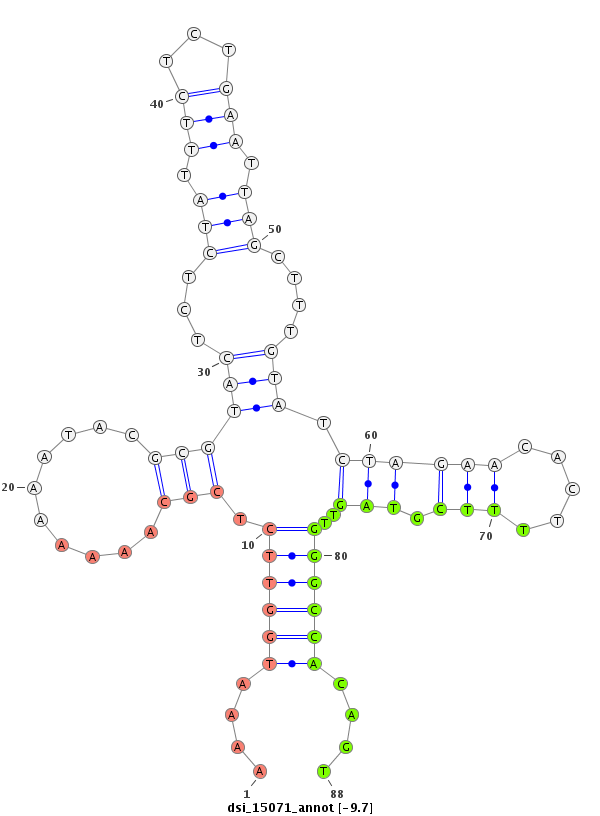 |
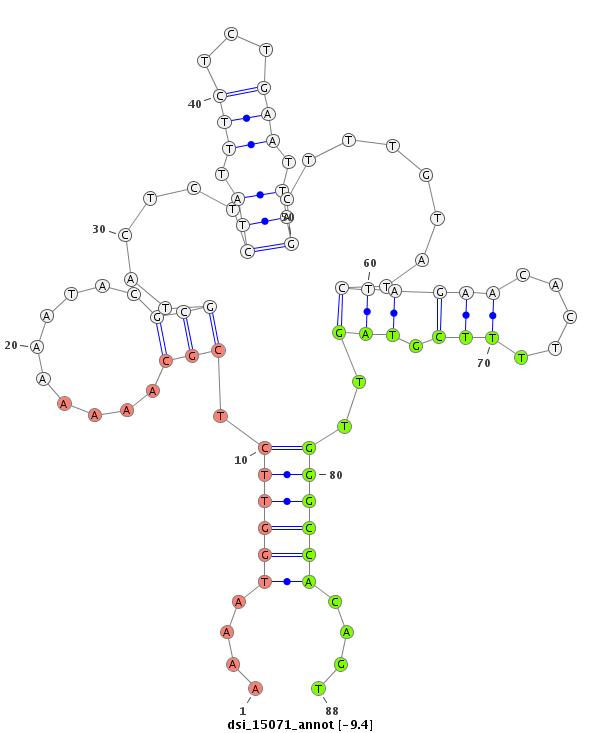 |
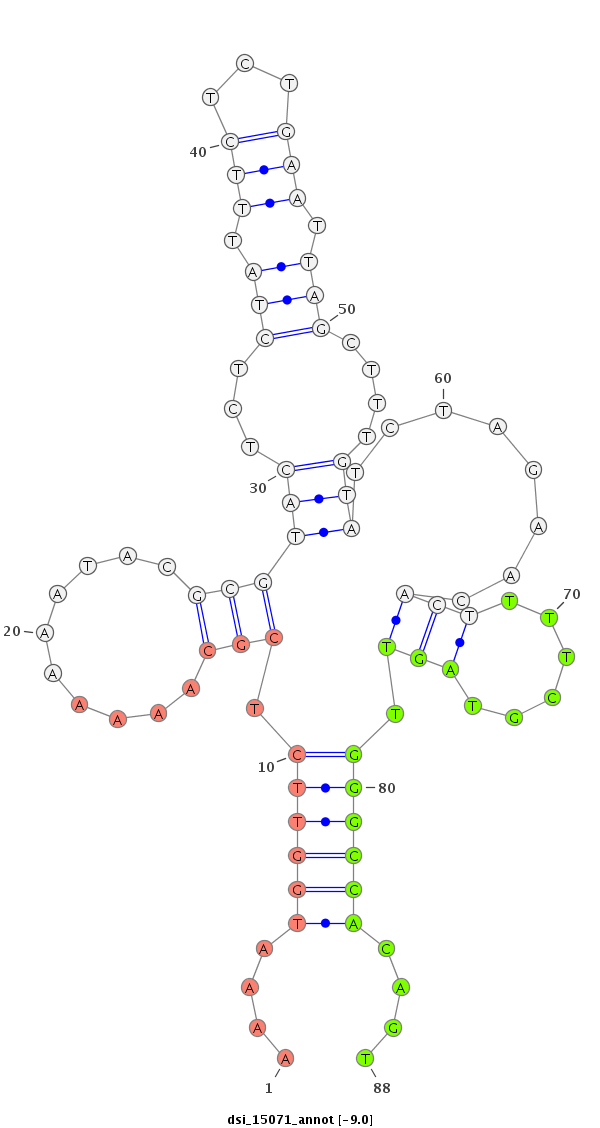 |
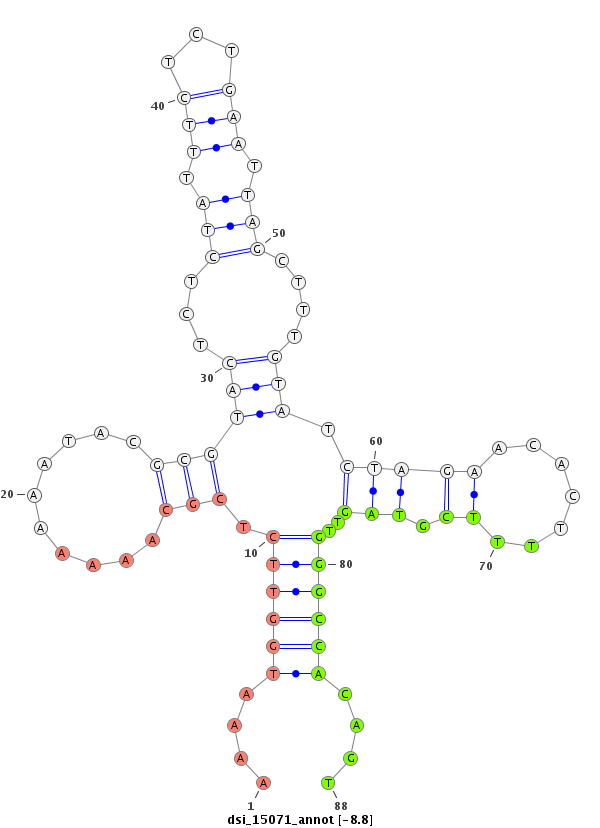 |
exon [3r_10116800_10116890_-]; CDS [3r_10116800_10116890_-]; intron [3r_10116891_10118002_-]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AATTCGTTGAAAATAGCCAGCGCAAGATATCAAATAACGCACTGAAATTTTCGTTTGGCTTGCCAAAAAAAATGGTTCTCGCAAAAAAATACGCGTACTCTCTATTTCTCTGAATTAGCTTTGTATCTAGAACACTTTTCGTAGTTGGGCCACAGTGTTTCCAAAAAGAATTCGAATTGCGATTCGTCTTTTGATTTTAGGAAAGTATTTCAAGGACATTTGGCATCGTTTCAAAAAGATCACAGCAGGT ********************************************************************...((((((((((.............((((...(((.(((...))).)))....)))).............)).))...))))...))********************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
O001 Testis |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
M024 male body |
SRR902009 testis |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................TTTCGTAGTTGGGCCACAGT.............................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................AAAATGGTTCTCGCAAAA.................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................AGAACACTTTTCGTAGTTGGGC.................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................ATTGCGATTCGTCTTTTGATTTTAGGAA............................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................GAATTGCGCTTCGTCT............................................................. | 16 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................AAATTTTCGTTTGGCTT............................................................................................................................................................................................. | 17 | 0 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................CCAGGGCAAGATATC........................................................................................................................................................................................................................... | 15 | 1 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................ATTTTCGTTTGGCTT............................................................................................................................................................................................. | 15 | 0 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................TTTGGCTTGAGAAAAAAAA.................................................................................................................................................................................. | 19 | 2 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................TTTTCGGACAGTATTTCAAC.................................... | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................TTTCGTAGTGGGGACAC................................................................................................. | 17 | 2 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................CGTGTTTTGAGTTTAGAAAA.............................................. | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................................TCGGCATCGTTTTAAAAAAA........... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TTAAGCAACTTTTATCGGTCGCGTTCTATAGTTTATTGCGTGACTTTAAAAGCAAACCGAACGGTTTTTTTTACCAAGAGCGTTTTTTTATGCGCATGAGAGATAAAGAGACTTAATCGAAACATAGATCTTGTGAAAAGCATCAACCCGGTGTCACAAAGGTTTTTCTTAAGCTTAACGCTAAGCAGAAAACTAAAATCCTTTCATAAAGTTCCTGTAAACCGTAGCAAAGTTTTTCTAGTGTCGTCCA
**********************************************************************************************...((((((((((.............((((...(((.(((...))).)))....)))).............)).))...))))...))******************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM343915 embryo |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR902009 testis |
M023 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................TGTCACATAGTTTTGTCTTAAG............................................................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................AGCCGGTGTCACAAAGG........................................................................................ | 17 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................TGCGCATGACAGATAA................................................................................................................................................ | 16 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................TAGCAAAGTTTTCCTA.......... | 16 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................CCGTAGTCAAGTTTTTCT........... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........TTTATCGGTCGCGTTAGAGA............................................................................................................................................................................................................................ | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................TTCAGATTAACACTAAGCAG.............................................................. | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................................................................................................................................................................................CGCAGAAAAGTTTTTCGAGT........ | 20 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................ACCGTAGCAACGTTCTT............. | 17 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................ACAACTCGGTGTCGCAAAG......................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
Generated: 04/24/2015 at 10:29 AM