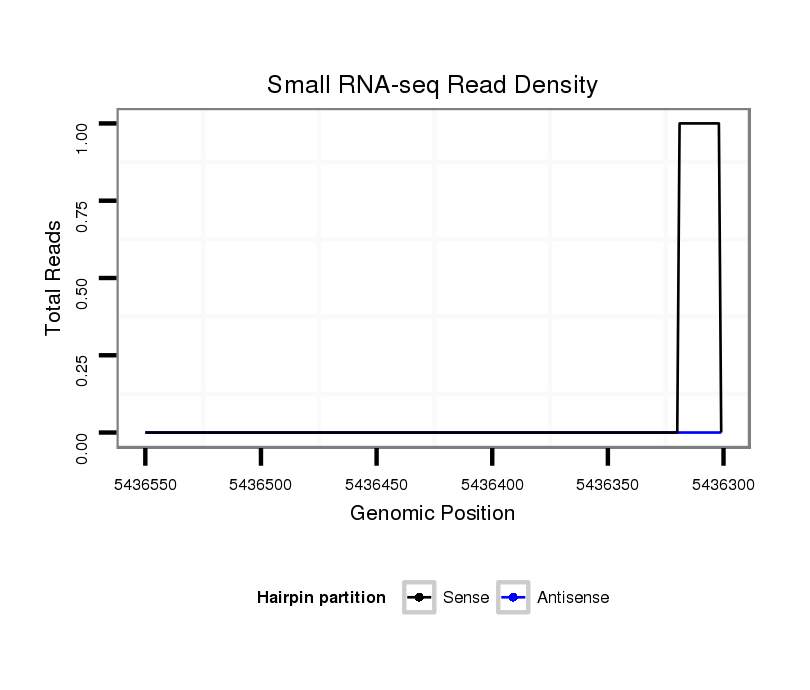
ID:dsi_14671 |
Coordinate:2r:5436351-5436500 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
CDS [2r_5436501_5436585_-]; exon [2r_5436501_5437013_-]; intron [2r_5435525_5436500_-]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ACTGCCTGAGTTCTTCGACAGCTTCGTGCAAAAGTGCACGGATCCCAGTTGTAAGTACATATGATGAAGGAGATTCTCTCTAAAAGTAACAATATTTATAGAAATATATATAAATAAGATTTAATGCAATCATGTTAGAAATTTTAAATGTTATAATAGTTTTAAGGTTGAGTACGGTTTTTAGTCAATGTAGTTTATATATAACTTTTTAACTATTATAACTTTTTAACTTTTAATAAGTAAATAAACG **************************************************((((((((((.(((((((((..((((.((.(((((((((((.((((((((........))))))))((((((...(((....)))...))))))....)))))).....))))).))..))))....)))))).)))))))).)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
SRR902009 testis |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................TTTAATAAGTAAATAAAC. | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................TGAGTACGGTATCTAGTGAAT............................................................. | 21 | 3 | 3 | 0.67 | 2 | 0 | 2 | 0 | 0 | 0 |
| ......................................................................................TAACCATATTTATAGAAATATTTAAA.......................................................................................................................................... | 26 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................................................TTTTGATCTTTTGATAAGTAAA...... | 22 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................TGAAGATTGAGTACGGT........................................................................ | 17 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................GATGAACGAGATTCCC............................................................................................................................................................................ | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
|
TGACGGACTCAAGAAGCTGTCGAAGCACGTTTTCACGTGCCTAGGGTCAACATTCATGTATACTACTTCCTCTAAGAGAGATTTTCATTGTTATAAATATCTTTATATATATTTATTCTAAATTACGTTAGTACAATCTTTAAAATTTACAATATTATCAAAATTCCAACTCATGCCAAAAATCAGTTACATCAAATATATATTGAAAAATTGATAATATTGAAAAATTGAAAATTATTCATTTATTTGC
**************************************************((((((((((.(((((((((..((((.((.(((((((((((.((((((((........))))))))((((((...(((....)))...))))))....)))))).....))))).))..))))....)))))).)))))))).)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
M025 embryo |
M023 head |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
M024 male body |
SRR553485 Chicharo_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................CAACACATCCCAAAAATCAGGTA............................................................. | 23 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................CTAATAGAGATTTTCGTTG................................................................................................................................................................ | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................TCAAAATTCGAACTCA............................................................................. | 16 | 1 | 10 | 0.40 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................AAAACTTGATAATATTTAATAATT..................... | 24 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........CAAGAAGCTGTCGAAGT................................................................................................................................................................................................................................ | 17 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................ATCAAAATTCGAACTC.............................................................................. | 16 | 1 | 12 | 0.33 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................TCAACATTGATGTATA............................................................................................................................................................................................ | 16 | 1 | 7 | 0.29 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................AAAATTCCAACTGATG........................................................................... | 16 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........TCAAGAAGTTGCCGAAGC................................................................................................................................................................................................................................ | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....GGACTCGAGAAGCTGCC..................................................................................................................................................................................................................................... | 17 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................AGATTATCATTGTTCTA........................................................................................................................................................... | 17 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
Generated: 04/23/2015 at 02:41 PM