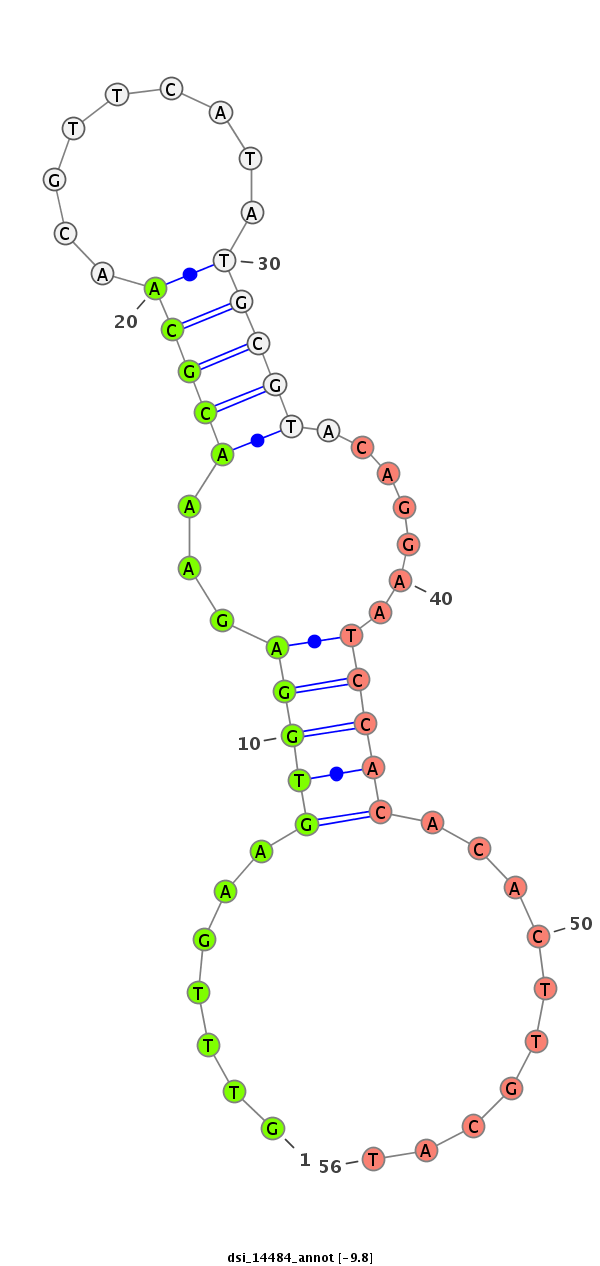
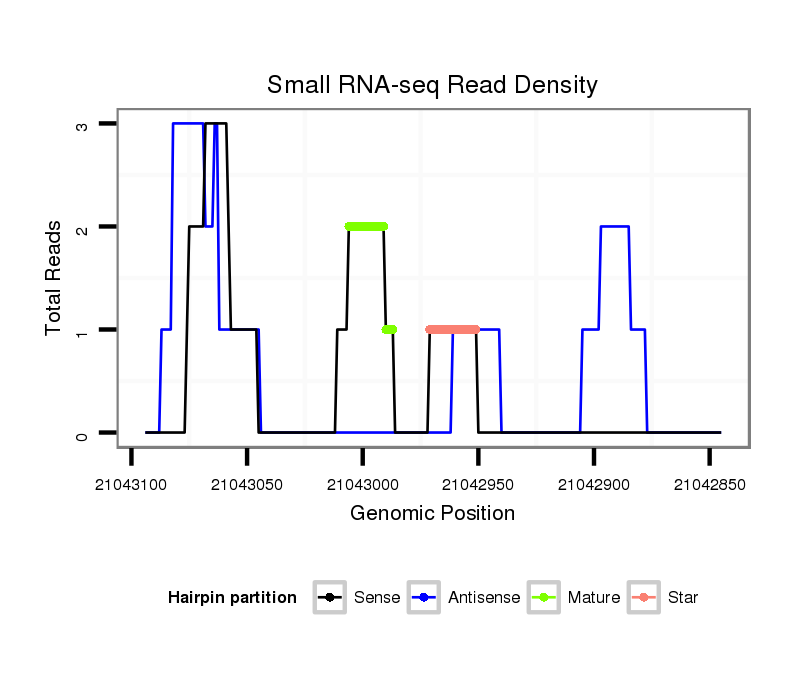
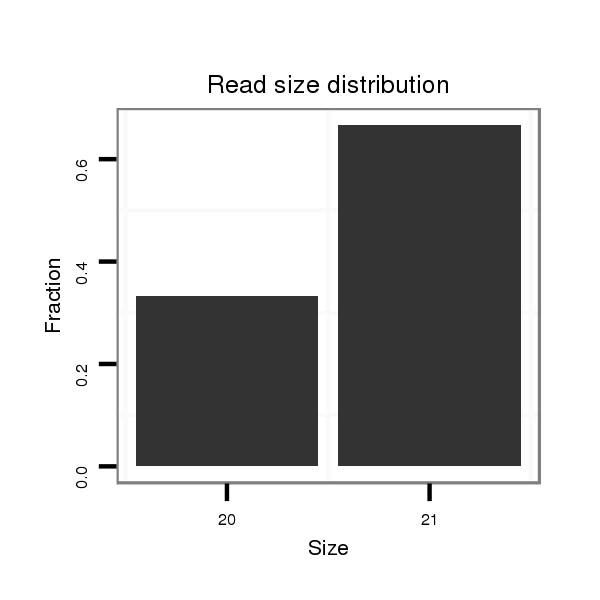
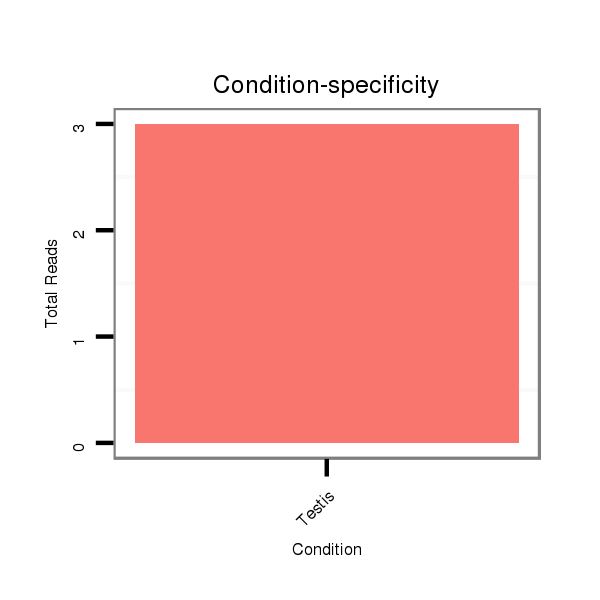
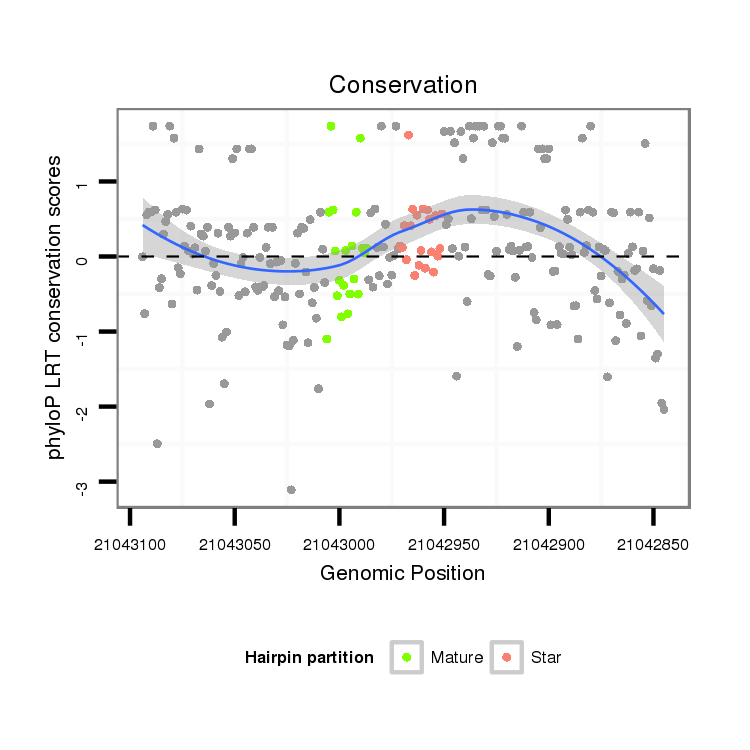
ID:dsi_14484 |
Coordinate:3l:21042895-21043044 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -9.8 | -9.0 | -9.0 |
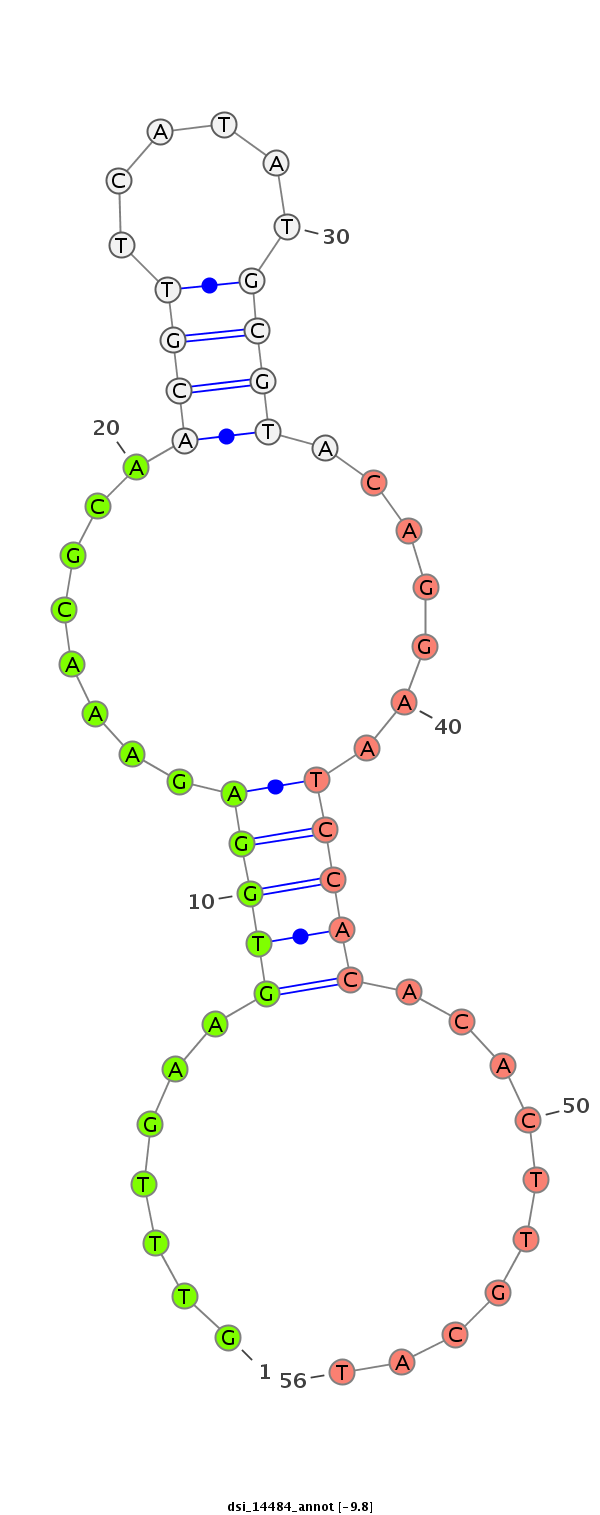 |
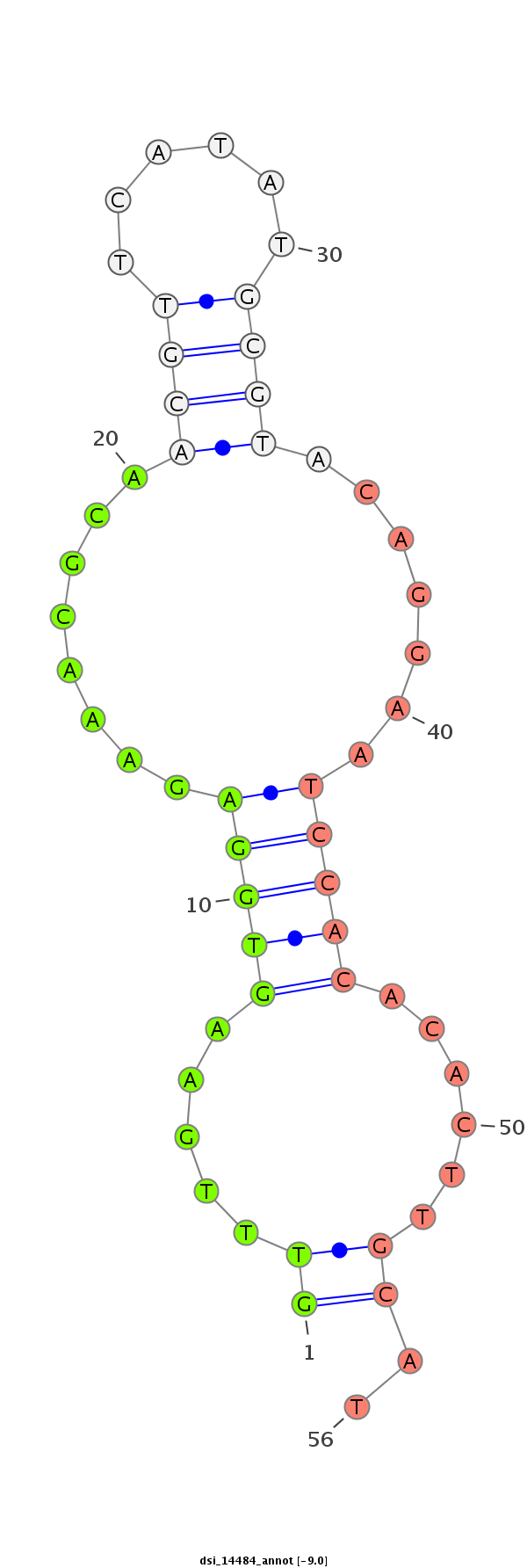 |
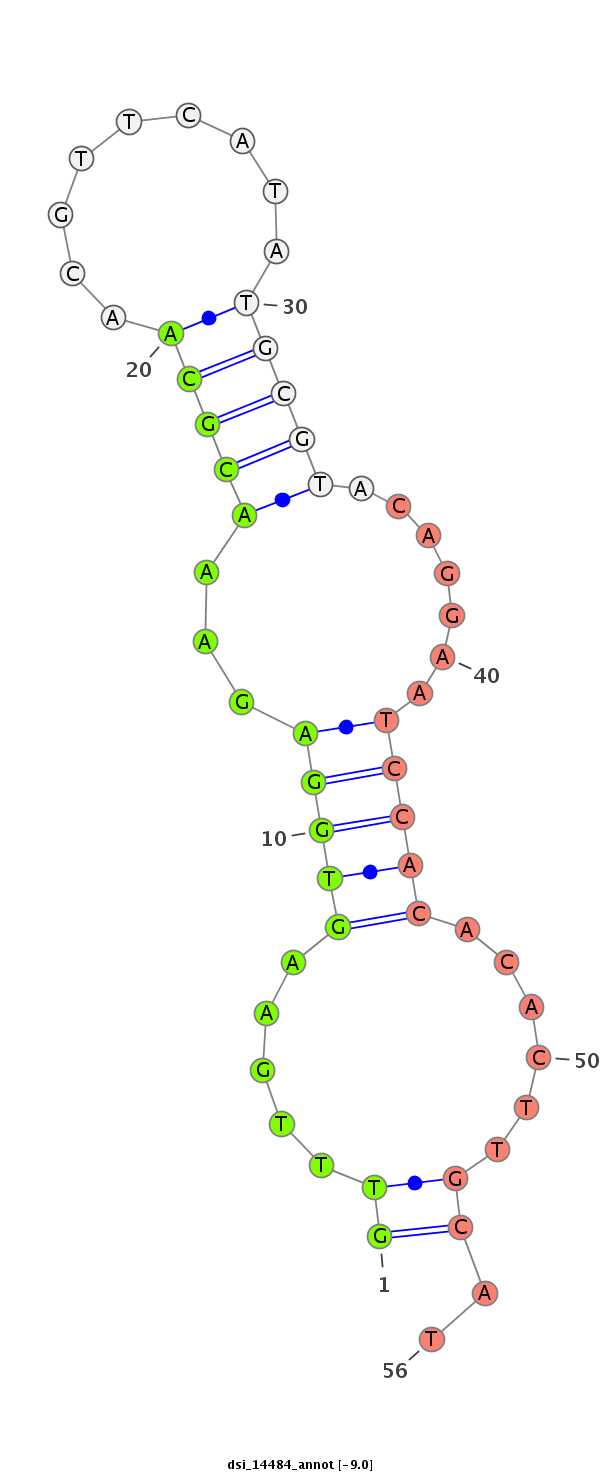 |
exon [3l_21043045_21043205_-]; five_prime_UTR [3l_21043045_21043205_-]; intron [3l_21042663_21043044_-]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CATTAAACCAGCTTCGATTTCAAGAAGTCGCACTCCCCGAAAGCCAGCAGGTAATGTAAAACATTCATCGCGTGCGATGCAAGCTTTAGTTTGAAGTGGAGAAACGCAACGTTCATATGCGTACAGGAATCCACACACTTGCATACATACTTACATTGCATATTTCACTGATGCTAAGGGGATAATTGAAATGCAGAAGTTGTCACGAGTGCATTTCGTGTGGTTTCCTGCTGAGGATTGCGAAAGCTCC ****************************************************************************************.......(((((...(((((.........))))).......)))))..........********************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
M023 head |
M025 embryo |
M024 male body |
O001 Testis |
M053 female body |
SRR553485 Chicharo_3 day-old ovaries |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................ACGCAACGTTCATCGGCGT................................................................................................................................ | 19 | 2 | 1 | 7.00 | 7 | 5 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................GCAACGTTCATCGGCGTA............................................................................................................................... | 18 | 2 | 3 | 3.33 | 10 | 5 | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CGCAACGTTCATCGGCGTAG.............................................................................................................................. | 20 | 3 | 2 | 2.50 | 5 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................ACGCAACGTTCATCGGCGTAG.............................................................................................................................. | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................ACGCAACGTTCATCGGCGTAGA............................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TAGATTGAAGTGGAGA.................................................................................................................................................... | 16 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................GTTTGAAGTGGAGAAACGCA.............................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................TTCAAGAAGTCGCACTCC...................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................ACGCAACGTTCATCGGCGTA............................................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CGCAACGTTCATCGGCGT................................................................................................................................ | 18 | 2 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CGCAACGTTCATTGGCGTA............................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CGCAACGTTCATCGGCGTA............................................................................................................................... | 19 | 2 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................CTTTAGTTTGAAGTGGAGAAA.................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................TCAAGAAGTCGCACTCCC..................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................GCAACGCTCATCGGCGTACAG............................................................................................................................ | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................CAGGAATCCACACACTTGCAT.......................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................GTCGCACTCCCCGAAAGCCAGCA......................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................CGTAACGTTCATCTGCGTA............................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................GCAACGTTCATCGGCGT................................................................................................................................ | 17 | 2 | 10 | 0.70 | 7 | 2 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................TTCCTGCTTGGGATTGTGAA...... | 20 | 3 | 14 | 0.57 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 |
| .......................................................................................................ACGCAACGTTCATCTGC.................................................................................................................................. | 17 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TGCTGAGGATTGCGAAAATT.. | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................TACGCAACGTTCATCGGCGTA............................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................CAACGTTCATCGGCGTAGAG............................................................................................................................ | 20 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CGCAACGTTCATCGGCG................................................................................................................................. | 17 | 2 | 11 | 0.45 | 5 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................GCAACGTTCATCGGCGTAG.............................................................................................................................. | 19 | 3 | 10 | 0.40 | 4 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................CAACGTTCATCGGCGTA............................................................................................................................... | 17 | 2 | 8 | 0.38 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................ACGCAACGTTCATCGGC.................................................................................................................................. | 17 | 2 | 12 | 0.25 | 3 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................ACGCAACGTTCATCGGCG................................................................................................................................. | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................TCGTGCTGGGGATTGCGA....... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CGCAACGTTCATCGGCGCA............................................................................................................................... | 19 | 3 | 17 | 0.24 | 4 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................................TCGTGCTGGGGATTGCG........ | 17 | 2 | 13 | 0.23 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................AGGCAACGTTCATAT.................................................................................................................................... | 15 | 1 | 9 | 0.22 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................TTTCATGCTTAGGATTGTGAA...... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................TACGCAACGTTCATCGGCGT................................................................................................................................ | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................TTTCATGCTTGGGATTGCGAA...... | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................................................CTGAGTATTGCGAAA..... | 15 | 1 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................ACGTAACGTTCATCGGCGT................................................................................................................................ | 19 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................CAAGCTTTTGTTTGAA........................................................................................................................................................... | 16 | 1 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................GATTGAAGTGGAGAA................................................................................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................TTGAAATGCAGGAGT.................................................. | 15 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................TTTCATGCTTAGGATTGTG........ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................ATGTTTGAAGTGGAGAAA.................................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................CAACGTTCATCGGCGTAG.............................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................TATGGGTTCCTGATGAGGA............. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GTAATTTGGTCGAAGCTAAAGTTCTTCAGCGTGAGGGGCTTTCGGTCGTCCATTACATTTTGTAAGTAGCGCACGCTACGTTCGAAATCAAACTTCACCTCTTTGCGTTGCAAGTATACGCATGTCCTTAGGTGTGTGAACGTATGTATGAATGTAACGTATAAAGTGACTACGATTCCCCTATTAACTTTACGTCTTCAACAGTGCTCACGTAAAGCACACCAAAGGACGACTCCTAACGCTTTCGAGG
**********************************************************************************************************.......(((((...(((((.........))))).......)))))..........**************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
O001 Testis |
M024 male body |
GSM343915 embryo |
SRR618934 dsim w501 ovaries |
M023 head |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............AAGCTAAAGTTCTTCAGCGT.......................................................................................................................................................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................TTACGTCTTCAACAGTGCTCA........................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................GTGTGAACGTATGTATGAATG................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......GGTCGAAGCTAAAGTTCTT................................................................................................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TTGTCCTTAGGTGTGTGAACGTAT......................................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................TCAACAGTGCTCACGTAAAG................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................GTGAGGGGCTTTCGGTCGTC........................................................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................GGAATGTGACGTATAGAGTG.................................................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................ATTACATTTTGTAGGTAC..................................................................................................................................................................................... | 18 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................AAGGACGACTTCTAA........... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................CCATTACATGTTGGGAGT....................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................GGGGCTTGCGGTCGTTC....................................................................................................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................TGTTAATAGCGCACGCTTC........................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......AGGTCGAAGCTAAAG..................................................................................................................................................................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3l:21042845-21043094 - | dsi_14484 | CATTAAACCAGCTTCGATTTCAAGAAGTCGCACT-CCCCGAAAGCCAGCAGGTAATG-------TAAAACAT-TCATCGCGTGCGATGCAAGC-TTTAGTTTGAAG----------------------------------------------------TGGAGAAACGCAA----------CGTTCAT----ATGCGTACAGGAATCCACACACTTGCATA--------CATA----CTTACAT--TGCATATTTCACTGA-----TGCTAAGGGGATAATTGAAATGCAGAAGTTGTC--ACGAGTGCATTT----------------CGTGTGGTT-TCCT--GCTG-AGGATTGCGAAA--GCTCC |
| droSec2 | scaffold_11:1446193-1446442 - | CATTAAACCAGCTTCGATTTCAAGAAGTCGCACT-CCCCGAAAGCCAGCAGGTAATG-------TAAAACAT-TCATCGCGTGCGATGCAAGC-TTTAGTTTGAAG----------------------------------------------------TGGAGGAACGCAA----------CGTTCAT----ATGCGTACAGGAATCCACACACTTGCATA--------CATA----CTTACAT--TGCATATTTCACTGA-----TGCTAAGGGGATAATTGAAATGCAGAAGTTGTC--ACGAGTGCATTT----------------CGTGTGGTT-TCCT--GCTG-AGGATTGCGAAA--GCTCC | |
| dm3 | chr3L:21526467-21526716 - | CATTAAACCAGCTTCGATTTCAAGAAGTCGCACT-CCCTGAAAGCAAGCAGGTAATG-------TAAAACAT-TCATCGCGTGCGATGCAAGG-GTTATTTTGAAG----------------------------------------------------TGCGGGATCGCAA----------CGTTCAT----ATGCGTACAGGAATCCTCGCACTTGCATA--------CATA----CTTACAT--TGCATATTTCACTGA-----TGCTAAGGGGATATTTGAAATGCAAAAGGTGTC--ACGAGTGCATTT----------------CGTGTGCTT-TCCT--GCTA-AGGATTGCGGAA--ACTCC | |
| droEre2 | scaffold_4784:21179995-21180244 - | CATTAAACCAGCTTCGATTATAGGAAGTCGCAGT-CACTGGGAGCCAGCAGGTAATG-------TAAAACAT-TCAAGGAAGGCAATGCAAGC-GTTAATTTGAAG----------------------------------------------------TTGTAGAACGCAA----------CGTTCAT----AGGCGTATAGGAAACCACACACTGGCATA--------CATA----CTAACAA--TGCATATTTCGCTTA-----TGCTAAGGGGATAATTTAAATGCAGAAGTTGTC--ACGACTGCATTT----------------CGTGTGGTT-TCCT--GCTG-AGGATGGCGAAA--ACAGC | |
| droYak3 | 3L:18989598-18989847 + | CATTAAACCAGCTTCGATTTTAGGAAGTCGCATT-CACTGAGAGCCAGCAGGTAATG-------TAAAACAT-TCAAGGAGGGCGATGCAAGC-GTTAGTTTGAAG----------------------------------------------------ATCAAGAACGCAA----------CGTTTAT----ACGCGTATAGGAATCCACACACTTGCATA--------CATA----CAAACAC--TGCATATTTCGCTGA-----TGCTAAGGGGATAATTTAAATGCAGAAGTTGTC--ACTAGTGCATTT----------------CGTATGGTT-TCCT--GCTG-AGGATTGCGAAA--ACTGC | |
| droEug1 | scf7180000409472:345943-346200 - | CGTTAAAG-AGCTTAGATTTTAGGAACTCGCGGA-GCCGTTAGGCCATCAGGTAATA-------TTGAACAGCTTACAATCAGCGATGAAAGTGTTTGATTTAATA----------------------------------------------------TCCAACAACAAAACTG----CTT---AAAT----ACGTCTATAGGAATTCACATACATATGTA--------CATGCGTACTTACAT--TGCATATTTTACTGA-----TGCTAAGGGAATTATACAAATGCATTTCTCGTC--ACGCTTGCATTT----------------CTGTTAAAT-TTCC--GCTG-AAAATAGCGAAG--AATTG | |
| droBia1 | scf7180000302377:1800170-1800430 + | CATTAAAATTGCTTTGATTTTAGGCAGTCCCAGT-GTCGTTCACCCAGCAAGTAATG-------CAAATCAG-CT-----TTGCTGTGGATGCGCTTGATTTGTTG----------------------------------------------------TCCGGGAGCACAACGC----CAG---AGAC----ACTTCTATAGGAATCCGCAAACATACACACATTTGCACATA----CCTACTT--TGCATACTCTTCTGATGCGATGCTACGCAAATAATTTAAATGCAAGACTGTTC--TCGCCTGCTTTT----------------CTCGTGATT-TCC---ACTATAAAATAG-TAAA--GCTTT | |
| droTak1 | scf7180000415388:207319-207589 - | CATTAATTCTGCTTGGATTTCAGGTAATCGCAGC-GTCGTTGAGCCAGCAGGTAATA-------TAAAATAG-CATTTAGCGGCGATAGACGCGTTTGATTCGATA----------------------------------------------------TCCCAGAACACACTTTCATA---CCTACATATGTACTTCTATAGGAATCCTCACGCATACATA----TGTACATT----CCTACAT--TGCATATTTCTCTGATGCGATGCTAGGGCAATAATTTGAATGCAAGAGTGTTC--ACGCCTGCATTT----------------CATGTGCTT-TTTC--ACCA-AAATTAGCAACA--AATCA | |
| droEle1 | scf7180000491255:2760379-2760624 + | GGTTAAATCATCTTTGAATTTAGGTAATTGTATT-GTTGTAGAGCCAGATAGTAGTA-------TATAAAAT-TTAAATATTGCGAGAAATTCATTAGATTTGGCA----------------------------------------------------TCCAAGAACACAGAGC----CAT---ACAT----ACACCTATAGGAATCTACATACATA--------------------CATACAA--TGCATATTTCGCTGA-----TGCTAAGGAAATAATTTGAATGCAAGAGTCGTC--ATACCTGCATTT----------------TATGTGTAT-TTCC--ATCG-AAAATAGCGACA--CATAT | |
| droRho1 | scf7180000779324:29724-29973 + | GGTTAAAATAGCTTAGATTTTTGGTAATTGCAAT-GTCGCGGAGCCAAAAAGTAACA-------ATAAAAGT-TTAAAAACTACCATGGATTCGTTTGATTTGACA----------------------------------------------------TCCAAGAACACCGAGC----CAT---ACAT----ACGCCTTTAGAAATCTACATAC----ATA--------CATA----CCTACAT--TGCATATTTCGCTGA-----TGCTAAGGTAATAATTTGAATGCAAGAGTCGTC--ATGCCTGCATTT----------------CATGTGTGT-TGCC--ACCG-AAAGTAGCGAAT--AATAG | |
| droFic1 | scf7180000453831:354687-354939 - | CGTTCAAACTGTTTAGATCTTAGGTTATTGCAAC-ATCGCAGAGCCAGCAGGTAAAA-------TTGTAAAGCATATATAATGCGATGATTTCCCTTCATTTGACG----------------------------------------------------CCGAAGAACACAGAGC----CAC---ACAT----A--TGTACG----TCTATATACACATGTA--------CATA----CCTACATCATGCATATTTCGCGGA-----TGCTTAGGGAATAATTGTAATGCAGGGGTCGAC--ACTGCTGAGCTT----------------TATGGGTTT-TCTTTAATCG-AGTATGGCAAAA--TTGGG | |
| droKik1 | scf7180000302694:396552-396815 - | GTTAAAA--AGCTTAGGCATTAAGTAATTGTAATTGTCAGTAAGCCATTGAATAAGGACTAAATTAATAAAT-TGGTAGCGCTTGATG----GGGCTCTCTTAGTA----------------------------------------------------CCCAAGAACTTTAATC----CTT---ACAT-----CGCCTATAACAGTGTACATATCTACATA--------CATATCTACTGACTT-TTGCATATTTAGCTAA---GATGCTAAGAAAAAAAACGTAGTGCAAGAACCATTTTTTATATGCATTC----------------GATGCGTGTTTTCT--TCAG-TAATTTGCA-TA--GCACA | |
| droAna3 | scaffold_13337:17106942-17107194 + | GCCTAAACTT--ATAGAACTTAGCTG----C--------------------------------------------------------ATATGCGCTCGTTTTGTC-CACAATAATCCGGCCCTAGATGGGCACCTGTGTAAGTGACTTTTCAAGGCTAACTAATTGCACAGATC----CTT---GCAA----ATGCCTTTTGGAGTTTATATACACACATA--------CATA---------AC--TACATATTTCG-TGA-----TGCTGAAGGCATTTT----------ATTTCGTC--ATGA-AGTATTTGCTAACCAAAAAGACGTGTTTAATA-CCTA--GTCG-AAGAACTCGATGGGGGTCT | |
| droBip1 | scf7180000396641:595669-595926 + | GCTTAAAGT--CTTAGAATTTAGCTG----C--------------------------------------------------------ATAGCTGCTTGATTTGTCTCGCAATAACCGGATACTAGATGGACACCTAGGTAACTGACTTTTCCAGGCAAACTAATTTCACAGATC----CTT---GCAA----TTGCCTATTGGAGTTTATATACACACATA--------TGTA----CATA-AT--TACATATTTCGC-GA-----TGCCAATAGCATTTT----------AAGTCGTC--ATGA-AGTATTTGCTAACCGAAAAGACGTGTTTAATA-CCTC--GCTG-AAGAACACGAAGGGGGTCT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
Generated: 05/18/2015 at 12:12 PM