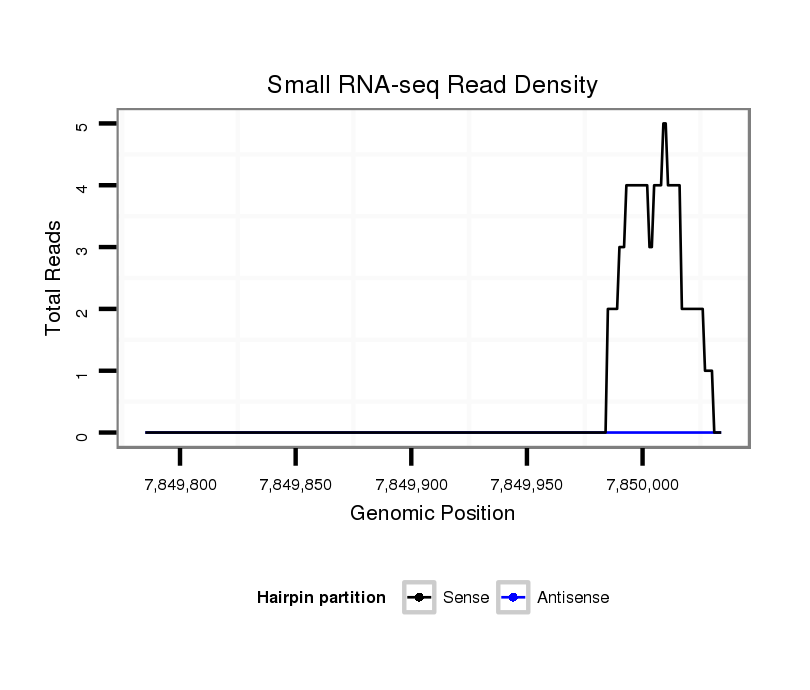
ID:dsi_14379 |
Coordinate:3l:7849835-7849984 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
intergenic
No Repeatable elements found
|
TGAGTGGCTGCTTTTCGGGCGTCGCAGCCGAAACTTTTGGGCTAAAAGCAAATGTAACTAATTGAAAAAGTTTTAAGCACAAACAAACAAGGCAAAAAAGATGTGTTGAAAGGGAGTTTCAAACTTTTGTTTGCTCCTTTCGCTCGTGTCGCTATAAAATTGGTTTCCACCTTAACTCATCTCTTCGAATCGATCAATAGCTATGTGATAACGGAGCTGCTGCAATCGGATCTGCACAAGATAATCGTGT
**************************************************.............((((.(((((((((((((......(((.(((.......))).))).(((((((...(((((....))))))))))))......))).))).)))))))..)))).................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
SRR553487 NRT_0-2 hours eggs |
M025 embryo |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR902009 testis |
SRR553488 RT_0-2 hours eggs |
M053 female body |
M023 head |
SRR553485 Chicharo_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................TAACGGAGCTGCTGCAATCGGATC.................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................ATCGGATCTGCACAAGATAATC.... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................CTATGTGATAACGGAGCT................................ | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................CTATGTGATAACGGAGCTGCTGCAAT........................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................TGATAACGGAGCTGCTGCAATCGGATC.................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................AAAGCTGTGTTGAGAGGGA....................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................TGCAATCGGATCTGCACAAGAT........ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................TGAGTCGCAGCCGAAACGTTT.................................................................................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................AGGTGTTGAAAGAGCGTTTCA................................................................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................TCCTTTCGCTCGTGCC.................................................................................................... | 16 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................AGCCAAAGCTTTTGGGCT............................................................................................................................................................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................AATCTACGTGATAACGGA................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................AAAAGCTGGGTTGAGAGGG........................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
ACTCACCGACGAAAAGCCCGCAGCGTCGGCTTTGAAAACCCGATTTTCGTTTACATTGATTAACTTTTTCAAAATTCGTGTTTGTTTGTTCCGTTTTTTCTACACAACTTTCCCTCAAAGTTTGAAAACAAACGAGGAAAGCGAGCACAGCGATATTTTAACCAAAGGTGGAATTGAGTAGAGAAGCTTAGCTAGTTATCGATACACTATTGCCTCGACGACGTTAGCCTAGACGTGTTCTATTAGCACA
**************************************************.............((((.(((((((((((((......(((.(((.......))).))).(((((((...(((((....))))))))))))......))).))).)))))))..)))).................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................GTAGAGGATCTTAGCTTGT...................................................... | 19 | 3 | 15 | 0.27 | 4 | 4 | 0 | 0 |
| .................................................................................................................................................................................GTAGAGAATCCTAGCTGGTT..................................................... | 20 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| .............................................................................................................................................................................TTAAGTATAGAAGCTTAG........................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 1 | 0 |
| ...........AAAGGCCCGCATCGTCGGCG........................................................................................................................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................AGACTATTGCCTCGA................................ | 15 | 1 | 15 | 0.07 | 1 | 0 | 0 | 1 |
Generated: 04/24/2015 at 10:30 AM