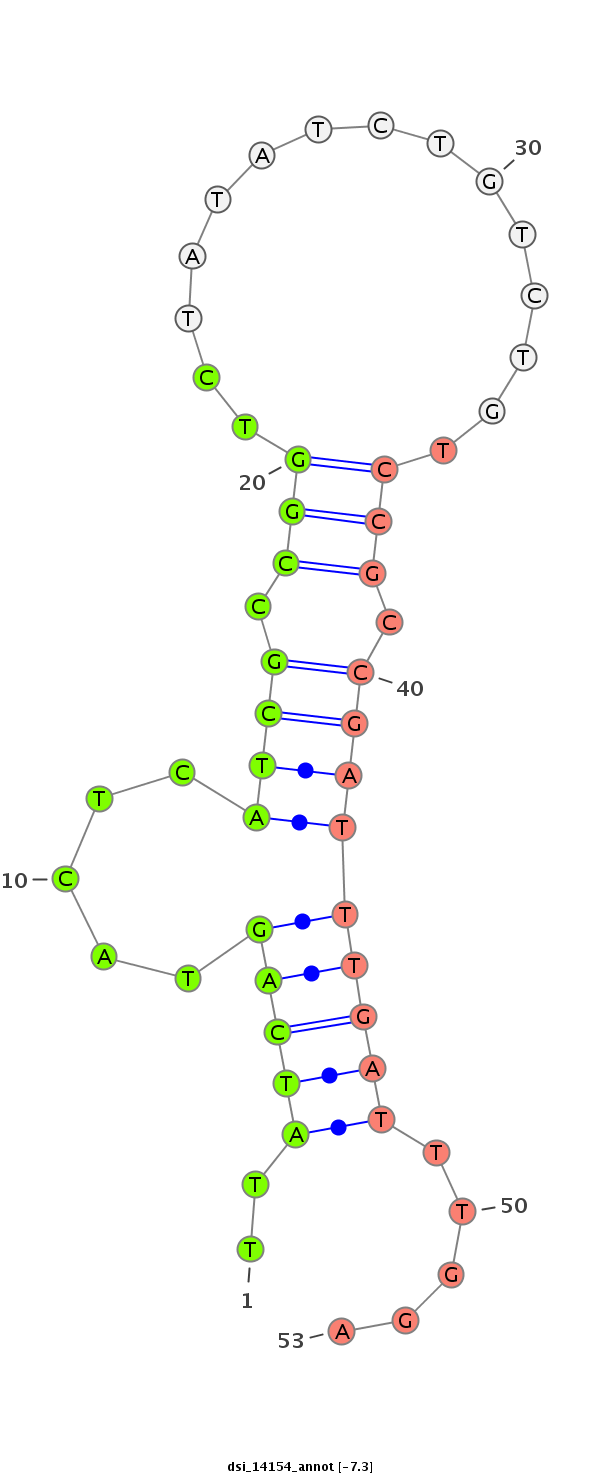
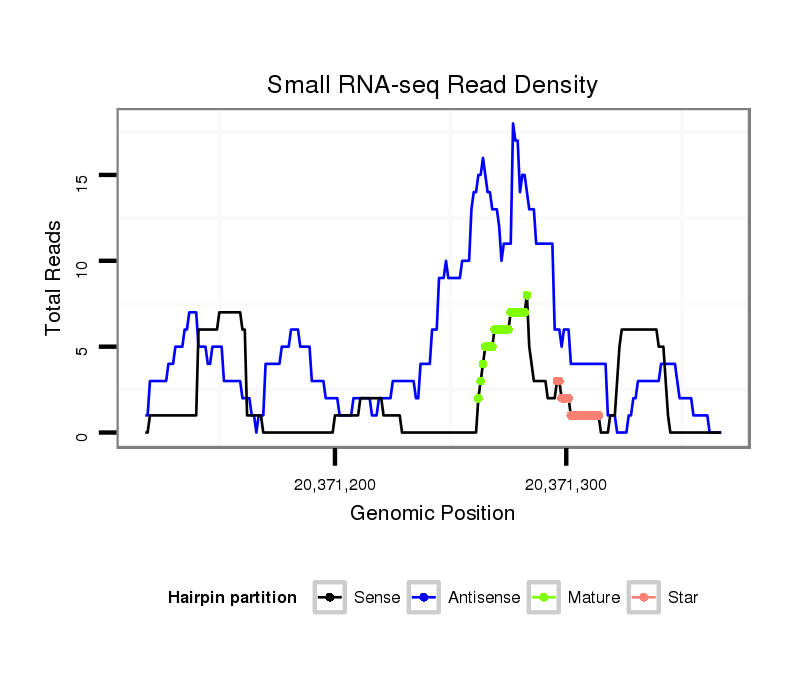
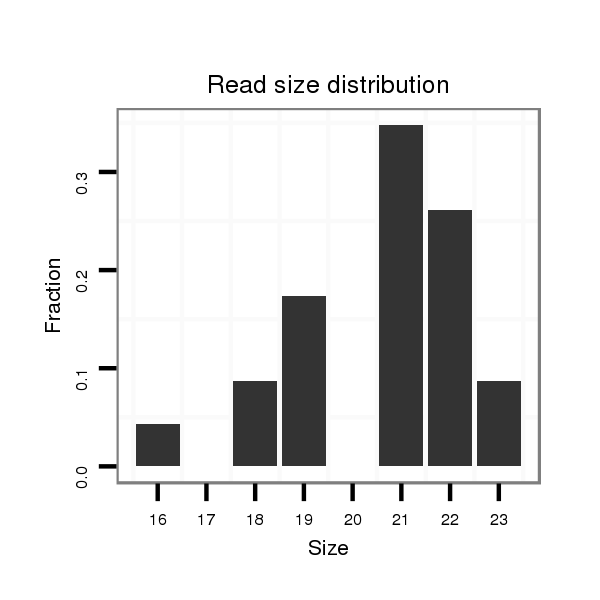
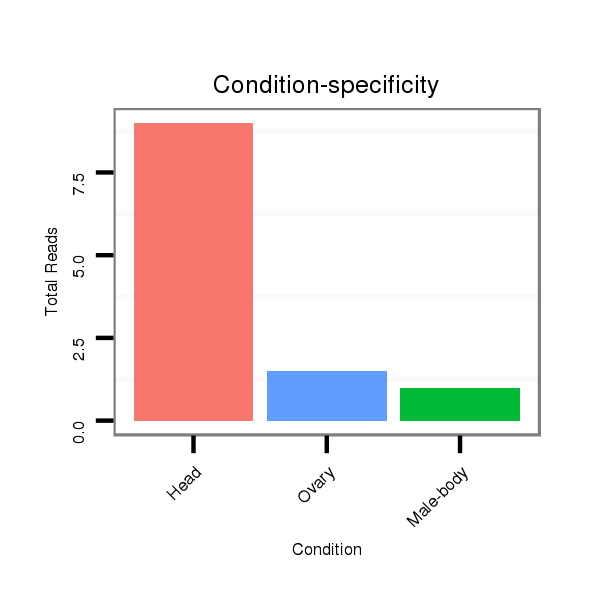
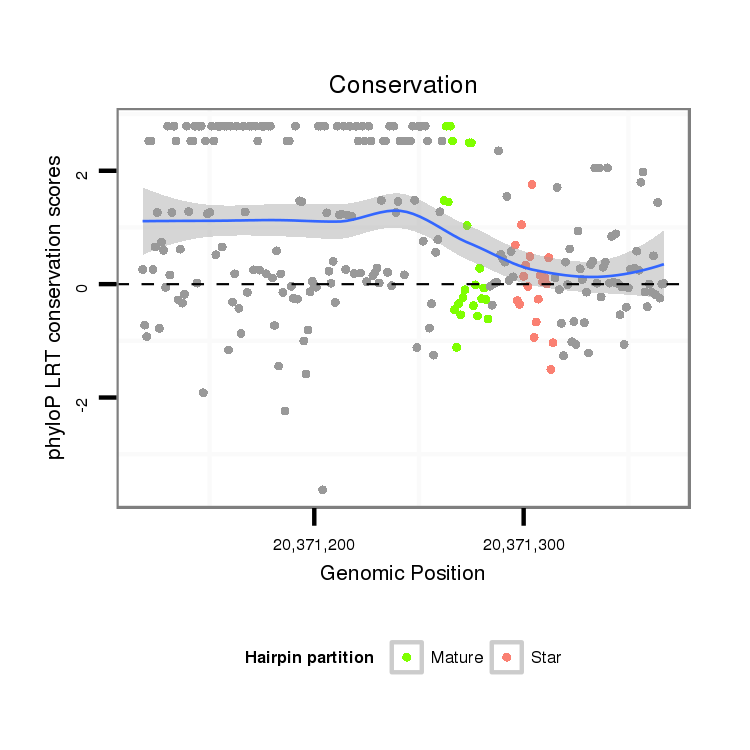
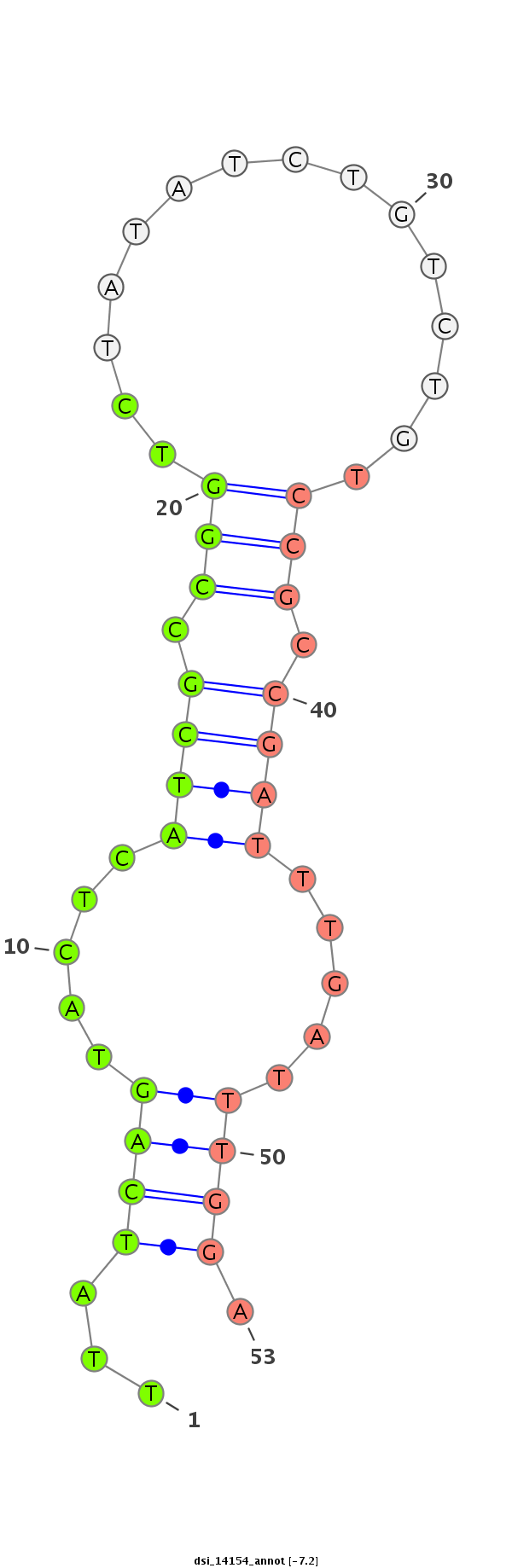
ID:dsi_14154 |
Coordinate:x:20371168-20371317 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -7.2 | -6.4 |
 |
 |
Antisense to five_prime_UTR [x_20371276_20371523_-]; Antisense to CDS [x_20370572_20371275_-]; Antisense to exon [x_20370572_20371523_-]
No Repeatable elements found
|
TGAGCCTCACTCAGCTGGCGGTCCCACTTTCGCTGGTTGTAGAGCAATAGGTAGTGGAAGTTCTTAACCCCGAACTTAACTTTGTATAGATCGAGCATTTCGTCCACTTCGCTATCGCTAAGTGGCCGCAAGATATCATCGTTCTTATCAGTACTCATCGCCGGTCTATATCTGTCTGTCCGCCGATTTGATTTGGATTTGGTTTTTCTGAACCTTCCGCCTGCCCGCTGCCAAAAGACTAAAGAGCTTG
************************************************************************************************************************************************..(((((.....((((.(((...............))).))))))))).....***************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M024 male body |
O002 Head |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M025 embryo |
O001 Testis |
SRR553485 Chicharo_3 day-old ovaries |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................CCACTTTCGCTGGTTGTAGAG.............................................................................................................................................................................................................. | 21 | 0 | 1 | 5.00 | 5 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................TTCTGAACCTTCCGCCTGCCC........................ | 21 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................TTTCTGAACCTTCCGCCTGCC......................... | 21 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TTATCAGTACTCATCGCCGGTC.................................................................................... | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................CCTGACGCCTGCCCGGTGCC.................. | 20 | 3 | 3 | 1.67 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................ATCAGTACTCATCGCCGGTCT................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................CTATATCTGTCTGTCCGCC.................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TTATCAGTACTCATCGCCT....................................................................................... | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................ATCAGTACTCATCGCCGGTCC................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................GTAGATCAATAGGTAG.................................................................................................................................................................................................... | 16 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................TACTCATCGCCGGTCTATATCTG............................................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................CGCCGGTCTATATCTGTCTGTC...................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................TATCAGTACTCATCGCCGGTC.................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CTGGTTGTAGAGCAATAGG....................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................TCCGCCGATTTGATTTGGA..................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................CCACTTTCGCTGGTTGTAG................................................................................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................TGTATAGATCGAGCATTTCGT................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................................TCTGAACCTTCCGCCTGCCCG....................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................TTTTTGAACCTTCCGCCT............................ | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................TCAGTACTCATCGCCGGTCTA.................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..AGCCTCACTCAGCTGGCGGTC................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GTTTTTCTGAACCTTCCGCCT............................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................AGCATTTCGTCCACTTCG........................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................GCCTGCCCGGTGCCGGAAGA............ | 20 | 3 | 3 | 0.67 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................GCCTGCCCGGTGCCGGAAG............. | 19 | 3 | 9 | 0.56 | 5 | 1 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................TTGTAGAGCAATAGGTATTA.................................................................................................................................................................................................. | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................ATCGTTCTTATCAGTA................................................................................................. | 16 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................CGCCTGCCCGGTGCCGGAAG............. | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................ACTCCGCTATCGCTACG................................................................................................................................ | 17 | 2 | 7 | 0.29 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................CGCCTGCCCGGTGCC.................. | 15 | 1 | 7 | 0.29 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................CCTGACGCCTGCCCGGTGC................... | 19 | 3 | 7 | 0.29 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATCTCTCTGTCCGCCG................................................................. | 16 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................TCGCTGGGTGTAGACC............................................................................................................................................................................................................. | 16 | 2 | 20 | 0.25 | 5 | 0 | 0 | 0 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................TTGATTTGGATTTGGGTTTT........................................... | 20 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................CTTATCAGTACTAATCGT......................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................CCTGCCCGGTGCCGGAAGA............ | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................GATATCATCGTTCTTGGC..................................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................CTGACGCCTGCCCGCTGC................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................CTGATCAGTACTAATCGCT........................................................................................ | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................ATTTGATTTGGAGATGGGTTT............................................ | 21 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................CAGTACTAATCGCTGG...................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................GCCTGCCCGGTGCCGGAA.............. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................GCCGATTTGATTGGGATTAA................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ########################################################################################################################################################################################################################################################## ACTCGGAGTGAGTCGACCGCCAGGGTGAAAGCGACCAACATCTCGTTATCCATCACCTTCAAGAATTGGGGCTTGAATTGAAACATATCTAGCTCGTAAAGCAGGTGAAGCGATAGCGATTCACCGGCGTTCTATAGTAGCAAGAATAGTCATGAGTAGCGGCCAGATATAGACAGACAGGCGGCTAAACTAAACCTAAACCAAAAAGACTTGGAAGGCGGACGGGCGACGGTTTTCTGATTTCTCGAAC *****************************************************..(((((.....((((.(((...............))).))))))))).....************************************************************************************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M024 male body |
M053 female body |
SRR553488 RT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
M025 embryo |
SRR902009 testis |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR902008 ovaries |
GSM343915 embryo |
O002 Head |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................CGGCCAGATATAGACAGA......................................................................... | 18 | 0 | 1 | 5.00 | 5 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................AGTCACGAGCACCGGCCAGA................................................................................... | 20 | 3 | 1 | 4.00 | 4 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................AAGAATAGTCATGAGTAGCGG........................................................................................ | 21 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TCACCTTCAAGAATTGGGGC.................................................................................................................................................................................. | 20 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................GCGGCCAGATATAGACAGGCA....................................................................... | 21 | 1 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................GTAGCGGCCAGATATAGACAGA......................................................................... | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................CGTTCTATAGTAGCAAGAATAGTCATG................................................................................................ | 27 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TCGAGGATTGGGGCTTGAAT............................................................................................................................................................................ | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................AATAGTCATGAGTAGCGGCCAG.................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ACTCGGAGTGAGTCGACCGCCAG................................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................AGTCATGAGCACCGGCCAG.................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................TTGAAACATATCTAGCTCGTA........................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................AAGAATAGTCATGAGTAGC.......................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................CGGCGTTCTATAGTAGCAAGAATAGT.................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TCACCTTCAAGAATTGGGGCTTGAAT............................................................................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............CGACCGCCAGGGTGAAAGCGA........................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................CAAGAATTGGGGCTTGAA............................................................................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................CCAGGGTGAAAGCGACCAACATCTCGT............................................................................................................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................AGCTCGTAAAGCAGGTGAAGCGATAGC..................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................GGATATAGACAGACAGGCGGC................................................................. | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................CTTGGAAGGCGGACGGGCGAC.................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................TAGTCATGAGTAGCGGCCAGATA................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................AGCGACCAACATCTCGTTA.......................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................CCATCACCTTCAAGAATT....................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................AGAATAGTCATGAGTAGCGGCCAGATA................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................TAGCAAGAATAGTCATGAGTAG........................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................AATTGGGGCTTGAATTGAAAC...................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................TTCACCGGCGTTCTATAGTAGCAAGAATA...................................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................AGGCGATAGCGATGCACTGGC.......................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................TTCACCGGCGTTCTATAGTAG.............................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................AGTCACGAGTACCGGCCAG.................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................CGGCGTTCTATAGTAGCAAGAA........................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................CGGCCAGATATAGACAGAC........................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GAAGGCGGACGGGCGACGGTTTTC............. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................CGGCCAGATATAGACAGACAGGCGG.................................................................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................CAGGCGGCTAAACTAAACCTAAACCAA.............................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................AGCGGCCAGATGTAGACAGGC........................................................................ | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................CAAGAATAGTCATGAGTAGCGG........................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........AGTCGACCGCCAGGGTGAAAGCGA........................................................................................................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................AGGCGGCTAAACTAAACCTAAA.................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................AAGCGATAGCGATTCACCGGCGTT....................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................ATAGCTCGTAAAGCAGGTGAAGCGAT........................................................................................................................................ | 26 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................CGTTCTATAGTAGCAAGAATAGTCAT................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..TCGGAGTGAGTCGACCGCCAGGGTG............................................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................TGGAAGGCGGACGGGCGACG................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................CGGCCAGATATAGACAGACAG...................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TAGTCATGAGTAGCGGCCA..................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................TCTATAGTAGCAAGAATAGTCATGA............................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................CAGATATAGACAGACAGGCGG.................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................CACCGGCGTTCTATAGTAGCAAGAAT....................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................CGGCTAAACTAAACCTAAA.................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................CGCCAGGGTGAAAGCGACCAACATC................................................................................................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................GGAATAGTCATGAGTAGCGGCCAGATA................................................................................. | 27 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................CAGGCGGCTAAACTAAACCTAAA.................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..TCGGAGTGAGTCGACCGCCAG................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................GGGCGACGGTTTTCTGATTTC...... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................CAGGTGAAGCGATAGCGATT................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................CTATTGTGGCTAGAATAGTC................................................................................................... | 20 | 3 | 4 | 0.75 | 3 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................CGTCGGTTTTCTGATGTCACG... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................AAGTGAGGCGATAGCGATGC................................................................................................................................ | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................AGAATAGTTCTGAGTAGCGGCT...................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................AGTCACGAGCACCGGCCAG.................................................................................... | 19 | 3 | 9 | 0.33 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................TGAGGCGATAGCGATGC................................................................................................................................ | 17 | 2 | 7 | 0.29 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................ATGATTACGGGCCAGATATA............................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................TGGTATCCATCACCTTCGA............................................................................................................................................................................................ | 19 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................TGAAAGTTATCTAGCTCG.......................................................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................................................................................................................................AAAAAAACTTGTAAGGCGG............................. | 19 | 2 | 16 | 0.19 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................CGTCGGTTTTCTGATGTCAC.... | 20 | 3 | 12 | 0.17 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................AGCGATAGCGATGCA............................................................................................................................... | 15 | 1 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GGCGGGCGGGCGAGGGTT................ | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................GTCGGTTTTCTGATGTCACG... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................AATTCGGACAGGCGACGGTT................ | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................AAAAATACTAGGAAGGCG.............................. | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................GTCACGAGCACCGGCCAGA................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................TTGAATTGAAACATA................................................................................................................................................................... | 15 | 0 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................AGCTAGTAAAGCAGG................................................................................................................................................. | 15 | 1 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....GGAGTCAGTCGACCG....................................................................................................................................................................................................................................... | 15 | 1 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................................................................AGAACGCGGCCGGGCGACG................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................GAGCAACTTCTCGTTAT......................................................................................................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................AGGTGACGCGATAGCCAG.................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | x:20371118-20371367 + | dsi_14154 | TGAGCCTCACTCAGCTGGCGGTCCCACTTTCGCTGGTTGTAGAGCAATAGGTAGTGGAAGTTCTTAACCCCGAACTTAACTTTGTATAGATCGAGCATTTCGTCCACTTCGCTATCGCTAAGTGGCCGCAAGATATCATCGTT---------CTTATC---AGTACT------CATCGCCGGTCTA------------------------------------TA------------------------------TCTGTCT-----GTCCG-CCGATTTGATTTG----GATTTG-GTT-TT-----TCTGAACCTT--CCGCCTGCCCGCTGCCAAAAGACTAA--------------------AGAGCTTG |
| droSec2 | scaffold_55:7037-7286 + | TGAGCCTCGCTCAGCTGACGGTCCCACTTTCGCTGGTTGTAGAGCAATAGGTAGTGGAAGTTCTTAACCCCGAACTTAACTTTGTATAGATCGAGCATTTCGTCCACTTCGCTATCGCTAAGTGGCCGCAAGATATCATCGTT---------CTTATC---AGTACT------CATCGCCGGTTTA------------------------------------TA------------------------------TCTGTCT-----GTCCG-CCGATTTGATTTG----GATTTG-GTT-TT-----TGTGAACCTT--CCGCCTGCCCGCTGCCAAAAGACTAA--------------------AGAGCTTG | |
| dm3 | chrX:21850914-21851167 + | TGAGCCTCGCTCAGCTGGCGGTCCCATTTTCGCTGGTTGTACAGTAATAGGTAGTGGAAGTTCCTTATACCGAATTTAACTTTGTATAGATCAAGCAGTTCGTCCACTTCGCTATCGCTAAGTGGCCGCAAGATATCACCGTT---------CTTATC---TGAACT------CATTGCCGGTCTA------------------------------------TA------------------T--------GTATCTGTCT-----GTCAG-CCGATTCGATTTG----AATTTA-GTT-TT-----TGTGAACCTT--CCGCCTGCCCGCTGCTAAAAGACTAG--------------------AGAGCTGG | |
| droEre2 | scaffold_4690:18229050-18229300 + | TGAGCCTCGCGTAGCTGGCGATTCCACTTTCGCTGGTTGTACAGCAGCAGGTAGTGGAAGTTCTTTACCCCGAACTTAACTTTGTACAGATCTAGCAATTCGTCCACTTCGCTATCGCTAAGTGGCCGCAAGATATCACCGTC---------CTTATC---GCTACT------CATCGCCTGTCTA------------------------------------TA------------------------------TCTGTCT-----GTCCG-CCGATCCGATTTG----GATTTA-GTTTTG-----TGTGTGCCTT--CCGCCTGCCCACCGCCAAATGACTAG--------------------AGAGCTGG | |
| droYak3 | X:21296150-21296438 + | TGAGCCTCGCCTAGCTGGCGGTCCCACTTCCGCTGGTTGTACAGCAGTAGGTAGTGGAAGTTCTTTACCCCGAACTTGGCTTTGTACAGATCGAGCAGTTCGTCCACTTCGCTATCGCTAAGTGGCCGCAAGATATCACCGTC---------CTTATCGCCGCTACT------CATCGCCGGTCTAT---------------GGTCTATGATGTATGGTCTAT-GG------CCTGTGTATAC--------GTATCTGTCT-----GTCCG-CCAATTCGATTTG----GATTTA-GTT-TG-----TGTGTGCCTT--CCGCCTGCCTGCTGGCAAATGACTAG--------------------AGAGCCGG | |
| droEug1 | scf7180000407331:8650-8925 + | TTAGCCTCATCCAGCTGACAATCCCACTTTCGCTGGTTATACAGCAGCAGGTAATGGAAATTCTTTACACCATATTTAACTTTATAGAGATCGAGCAGTTCATCCACTTCACCGCCGGTAAGTGGCCGCAAAATATCACTGTC---------CTTATC---AAGGCC------CATGGTCAGACTAT------------------------------------A--------TGTATATGTATGTACATATGTATGTATGT-----ATTCG-TCAATTTTATCTGTGTCAACTTA-TAT-TG-----TATGTACTTT--CTGTATGCCCGCCGACTAATGACTGG--------------------CGAACTGG | |
| droBia1 | scf7180000302244:43309-43557 + | CCAGCCTCGTCCAACTGGCGGTCCCACTTCCGCTGGTTGTAGAGCAACAGGTAGTGGAAGTTCCTCACACCACACTTCTGTTGGTAAAGGCCGAGCAGTTCGTCCACCTCAGCGTCGCTGAGTGGCCGCAAGATATCCCCGGC---------CTTATC---GATGCT------CATCGCCGAGTCCT-----------------------------------AA------------------------------TCTGTCT-----GCCCGTCCGATCTGATCTG----GGTT----TT-TC-----GATGTGCTTT--CCGTCCGCCCGCCGTCCAGTGACTGA--------------------CGGGCCTG | |
| droTak1 | scf7180000415324:76094-76344 + | TTGGCCTCATCCAACTGGCGATCCCACTTTCGCTGGTTATAGAGCAACAGGTAGTGGAAGTTCTGTACACCATATTTAACTTTGTATAGATCGAGCAGTTCGTCTACTTCACTGTCGATAAGTGGCCGCAAAATATCACAGTC---------CTTATC---GATGCC------CATCGTCAAGCCCT----------------------------------ATA------------------T--------GTATCTGTCT-----AT---------CTGATTTG----TGTTTT-GTTTTA--TG-TATGTGCTTT--CCGTCTGCCCGCTGCCCAATGACTGG--------------------CGAGCTGG | |
| droEle1 | scf7180000491087:948835-949128 - | TGGGCCTCGTCCAACTGGGCATCCCACTTTCGCTGATTGTAGATCAGCAGGTAGTGGAAGTTCCTCACACCGAACTTGGATCTATACAGATCGAGCAGTTCGTCCACTTCACCATCGCTAAGTGGCCGCAAGATATCGCTATCAGTGTCCACCATATCATCGATGCC------CATCGCCAGATCCT----------------------------------AT-AACCAGTTATCACAGATAT--------GTATCTGTCT-----GTCTG-CCGAT-----TTT----AATTCA-CTT-TT-----TGTGTGGCTTCCCCGACTGCCTGCCGTCCGACGACTGGT--------CGAGGTTCATTAAAGCTGA | |
| droRho1 | scf7180000777745:44699-44945 - | TTGGCCTCGTCCAACTGTCGATCCCACTTGCGCTGATTGTAGATCAATAGGTAGTGGAAGTTCTTCACCCCATATTTGGCTTTGTACAGATCGAGCAGTTCGTCCACTTCACCATCGCTAAGTGGCCGCAAGATATCACCGTC---------CTTATC---GATGTC------CATATTCAAATCCT----------------------------------CTA------------------------------TCTGTCT-----GTCCG-CCGAT-----TTT----GCTTTA-TTC-TG-----CGTGTGGCTT--CCGCCTGCCCGACTTTTAATGACTGG--------------------CGAGCCGA | |
| droFic1 | scf7180000454116:104407-104656 + | CCGGCCTCGTCCAGCTGGCGATCCCACTTCCGCTGGTTGTACAGCAGCAGGTAGTGGAAGTTCCTCACCCCGAACTGGGCCTTATAGAGGTCGAGCAGCTCGTCCACCTCGCCATCGCTAAGTGGCCGCAGTATATCAGTGTC---------CTTATC---GCTGCC------CATCGCCAGAATCT----------------------------------CTA------------------------------TCCGTCT-----GTCCG-CCGAT-CGACTCG----GGTTTG-TTT-TG-----GGTTTA-GTT--CCGTCTGGCCGCCGTCCAATGACTGA--------------------CAAACTGG | |
| droKik1 | scf7180000302544:617944-618189 - | TGTGCCTCGTCCAGCTGGCGGTCCCATTTCCGCTGGTTGTACATCAACAAATAGTGGAAGTTCCTCACGCCGAACCTGGCCTTGTAGAGATCGAGCAGCTGGTCCACCTCGCTATCGCTGAGTGGCCGCAAGATATCGCCGACAAAGTCATCCTTATC---GCCATCGATTCCCATCGCGA--TCGAAATTGAG---------------------------------------------ATA------------------------------------CTACTGG----CTTTGA-GTG-TG-----TGTGTATGCT--CCGTCTGGCCGCTGTCCGGTGACTGA---------------------------- | |
| droAna3 | scaffold_13117:1965636-1965877 - | TTTGCTTCGTCTAGCTGGCGATCCCACTTTCTCTGATTGTAAAACAGCAGGTAGTGAAAGTTTTGGACTCCGAACTTGAGCTTGTAGAGATCGAGAAGCTCGTCTACTTCACTGTCGCTGAGTGGACGCAATATATCATTATC---------CTTGTC---GGTTCC------CATCGTCAGATCGT------------------------------------G--------TCCACATGTAC--------GTAT--------------------GTGTATT--------------------ATCGGGCTGTGTTTT--CTGTCTGGCCGCCTCCCAATCGCTAGA-------------------CAAACTGG | |
| droBip1 | scf7180000396425:101285-101526 + | TTGGCATCGTCCAACTGGCGATCCCACTTTCTCTGATTGTAAAACAGCAGATAGTGAAAGTTTTGGACTCCGAACTTGGTCTTGTACAGAGCTAGCAGCTCGTCTACTTCGCTATCGCTGAGTGGACGCAATATATCATTATC---------CTTGTC---GGTTCC------CATCGTCAGATCGT------------------------------------G--------ACCACATGTAC--------GTAT--------------------GTGTATT--------------------ATCGGGCTGTGTTTT--CTGTCTGGCCGCCGCTCAATCGCTAGA-------------------CAAACTGG | |
| dp5 | XL_group1a:1167374-1167630 - | CCCGCCGCCGCCAGTTGGCAGTCCCATTTGCGCTGATTGTAGAGCAGAAGATAGTGGAAATTCTGGGGCCCGTATTTCCGCTCGTAGAGACCGAGCAGCTCGTCGACCTCGTTATCGCTGAGTGGCCGCAAAATATCGATATC---------CTTATC---TGCATTGATGCCCAT------GCTG------------------------------------GT------------------------------TCTGTCACCTCTGTG----TGGTCTGGCTGG----GACT-GTT-GTGG-----TCTGATCGGA--CTGCCCGCTCGACTTTCAATGTCTCGATGACGGCTTGC---------------C | |
| droPer2 | scaffold_26:1282545-1282801 - | CCCGCCGCCGCCAGTTGGCAGTCCCATTTGCGCTGATTGTAGAGCAGAAGATAGTGGAAATTCTGGGGCCCGTATTTCCGCTCGTAGAGACCGAGCAGCTCGTCGACCTCGTTATCGCTGAGTGGCCGCAAAATATCGATATC---------CTTATC---TGCATTGATGCCCAT------GCTG------------------------------------GT------------------------------TCTGTCACCTCTGTG----TGGTCTGGCTGG----GACT-GTT-GTGG-----TCTGATCGGA--CTGCTCGCTCGACTTTCAATGTCTCGATGACGGCTTGC---------------C | |
| droWil2 | scf2_1100000004590:441917-442091 + | CCCGCCTTTTCTAGCTGTCTATCCCACTTCCGTTGATTGTAAAAGAGAAGATAATGAAAATTGAGTGGACCGTATTTTGATAGATAAAGCTGTAGCAGCTCATCCACTTCGCTGTCGTTTAATGGCCGCAATATATCCTTATC------TTGCTTATC---GGCCAT------TATTCTCAATCTC------------------------------------GA------------------------------TC--------------------------------------------------------------------------------------------------------------------- | |
| droVir3 | scaffold_12970:2606411-2606559 + | CCCGCCGCGTCCAGCTGTCTATCCCATTTGCGCTGATTGTACAGCAAAAGGTAGTGGAAATTGAGTGGTCCATATTTCTCGCCATACAACTGCATCAGCTCATCCACTTCGCTATCGCTGAGTGGTCGCAAAATATCGACATC---------CTTATC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droMoj3 | scaffold_6473:3333745-3333893 + | CCTGCTGCCTCAAGCTGGCGATCCCATTTACGCTGATTGTACATTAATAGATAGTGAAAGTTAAGCTCTCCATATTTCGCCTTATAGAGCGCCATCAGCTCATCCACTTCGATATCGCTGAGTGGCCGCAAGATGTCCAAATC---------CTTATC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15081:1174511-1174745 - | CCCGCCGCGTCCAGCTGGTGATCCCATTTGCGCTGATTGTACAGCAGCAGGTAATGGAAATTGAGCACACCATATTTGTCGCTATACAGCTGCATCAACTCGTCCACTTCGCTATCGCTTAGCGGACGCAAAATGTCGACATC---------CTTATC---ATTGAC------CATTTCTCGACTAGAGCTGCTAATTGCCG---C-----------------T--------ACA--ATGTCT--------GTGT----TT-----GTTT------------------------------GT--CGTCTGTCGTCTG--CCGTCTGCCTGTTGC--------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 01:35 PM