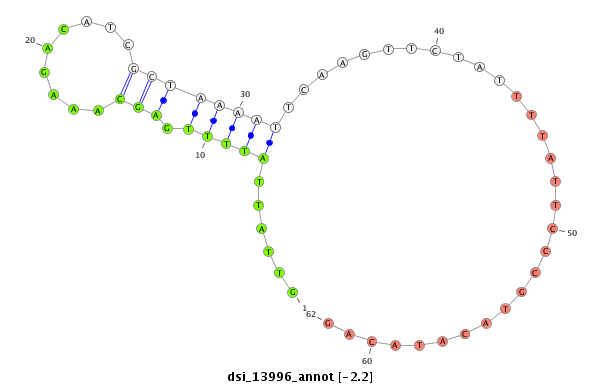
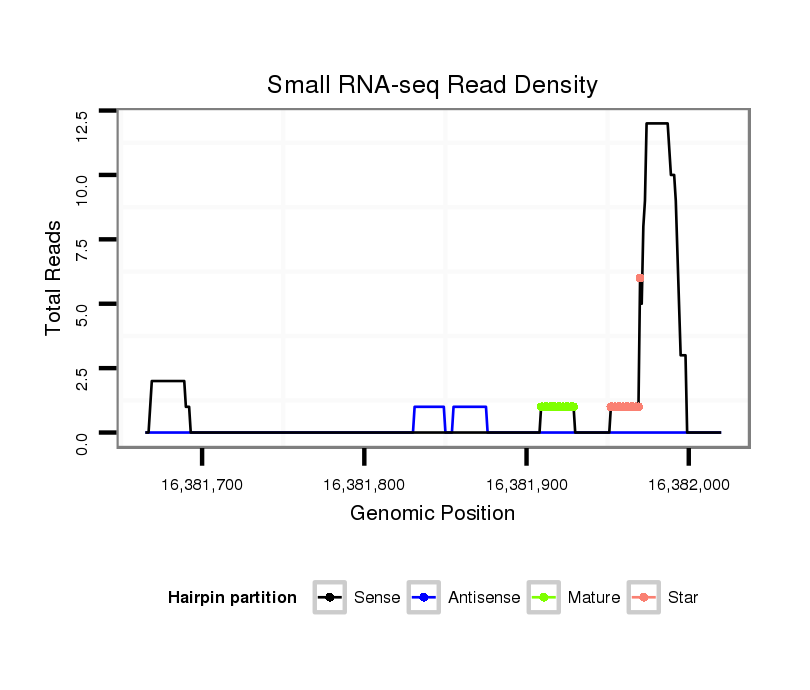

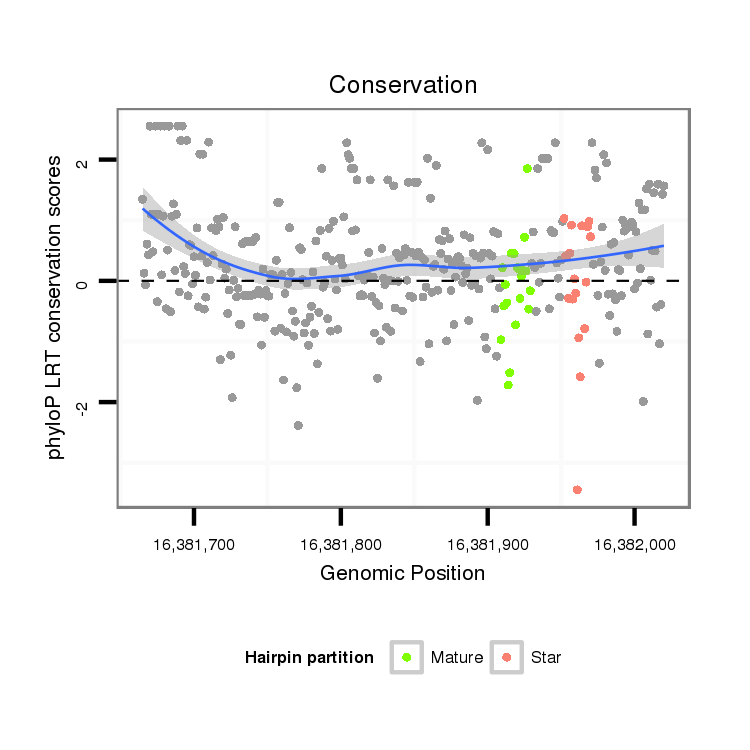
ID:dsi_13996 |
Coordinate:3r:16381715-16381970 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -1.9 | -1.2 |
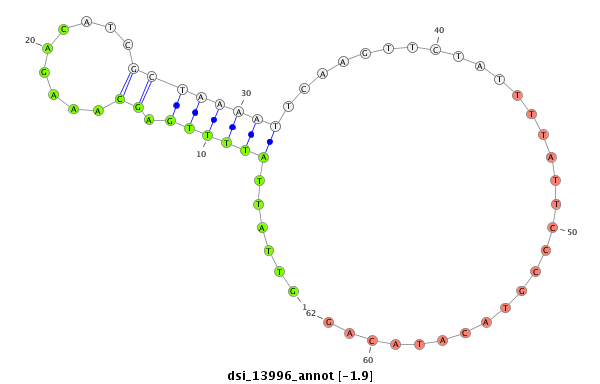 |
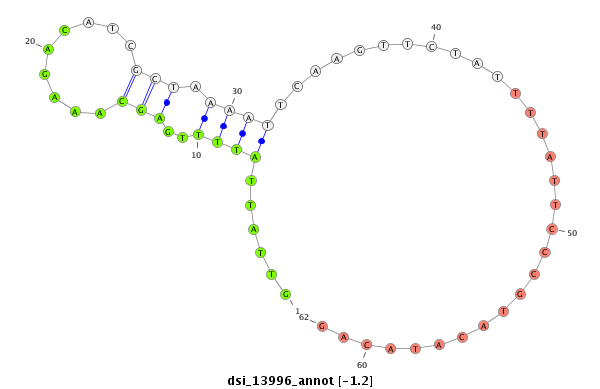 |
five_prime_UTR [3r_16381971_16383689_+]; exon [3r_16381971_16383689_+]; CDS [3r_16381971_16383689_+]; five_prime_UTR [3r_16381562_16381714_+]; exon [3r_16381562_16381714_+]; CDS [3r_16381562_16381714_+]; exon [3r_16381562_16381714_+]; exon [3r_16381971_16383689_+]; intron [3r_16381715_16381970_+]; intron [3r_16381715_16381970_+]
No Repeatable elements found
| ##################################################----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGCAGTGCGTCCAGCAACTGGATCTGGATGCCATAAAGCCAGGCGATCAGGTGTGTAATGCCCGCAGCCCTCAAGTTTCGGCTTTTGTTCGGTTTACATCTTACTAGAAACTCTACAGCACAGCTAGCACATTGATGTTTGTCAGTTATTCGTGGAAAAACTTACATACGTTGTTGCAAATATATACAATCCATACATTTATTGATTCCCAGATCACTATTTTCATTACCCTCGTTTAAACATTGTTATTATTTTGAGCAAAGACATCGCTAAAATTCAAGTTCTATTTTATTCCCGTACATACAGAACCGGACGGAAACTCCCAGAAGGCACAATCCAACACATAGTCACAGCCA ****************************************************************************************************************************************************************************************************************************************************......(((((.(((.........))))))))..............................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M024 male body |
SRR902009 testis |
M025 embryo |
M053 female body |
O001 Testis |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................................................................................................................................CGGACGGAAACTCCCAGAAGGCACA...................... | 25 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................ATCAGAACCGGACGGAAACTCC................................. | 22 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................GAACCGGACGGAAACTCCCAGAAG........................... | 24 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................ACCGGACGGAAACTCCCAGAA............................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................GAACCGGACGGAAACTCCCAGAAGG.......................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................CCGGACGGAAACTCCCAGAA............................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................ACCGGACGGAAACTCCCAGA............................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................GTTATTATTTTGAGCAAAGAC........................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....GTGCGTCCAGCAACTGGATCT........................................................................................................................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................TTTATTCCCGTACATACAG.................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...AGTGCGTCCAGCAACTGGATCTGGA........................................................................................................................................................................................................................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................GAACCGGACGGAAACTCCC................................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................ACCGGACGGAAACTCCCAGAAGG.......................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................GAACCGGACGGAAACTCC................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................GCAATCAGGTCTGTGATGCC...................................................................................................................................................................................................................................................................................................... | 20 | 3 | 7 | 0.29 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................ATGCGATAAAGCCAGTCG....................................................................................................................................................................................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................CAGAACCGGACGGAGA..................................... | 16 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| TGTAGTGCGTCCAGC..................................................................................................................................................................................................................................................................................................................................................... | 15 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................CCGGACGGAAACACCA................................ | 16 | 2 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| .............................................................................................................................................TAAGGTAATCGTGGAAAAA.................................................................................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................CGAACAGGTGAGTAATGC....................................................................................................................................................................................................................................................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................................................ACGGTGTTATTATTTTGA................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................GGCGAACGGGTGAGTAATG........................................................................................................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................CCTACACATAGTCAGA.... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................ATCGATGTTTGTCAGG.................................................................................................................................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
ACGTCACGCAGGTCGTTGACCTAGACCTACGGTATTTCGGTCCGCTAGTCCACACATTACGGGCGTCGGGAGTTCAAAGCCGAAAACAAGCCAAATGTAGAATGATCTTTGAGATGTCGTGTCGATCGTGTAACTACAAACAGTCAATAAGCACCTTTTTGAATGTATGCAACAACGTTTATATATGTTAGGTATGTAAATAACTAAGGGTCTAGTGATAAAAGTAATGGGAGCAAATTTGTAACAATAATAAAACTCGTTTCTGTAGCGATTTTAAGTTCAAGATAAAATAAGGGCATGTATGTCTTGGCCTGCCTTTGAGGGTCTTCCGTGTTAGGTTGTGTATCAGTGTCGGT
**************************************************......(((((.(((.........))))))))..............................**************************************************************************************************************************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
O001 Testis |
SRR553488 RT_0-2 hours eggs |
SRR902009 testis |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
M025 embryo |
M023 head |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................ATGCAACAACGTTTATATA........................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................TCTTCCGTATTAGGTTG............... | 17 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................CGATCTTTGAGAGGTCGTG........................................................................................................................................................................................................................................... | 19 | 2 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................GTGTTAGGTTGGTTATCAGT...... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................AAGTCCAAGATAAAGTGAGGG............................................................ | 21 | 3 | 2 | 1.00 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................GGTATGTAAATAACTAAGGGT................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................AGTGATAAAAGTAACGAGAGAA......................................................................................................................... | 22 | 3 | 5 | 0.80 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................TTGGGTCCGCTAGTCCAT............................................................................................................................................................................................................................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................GTGATAAAAGTAACGAGAGAAAA....................................................................................................................... | 23 | 3 | 6 | 0.50 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................ACAAGAACGCTTATATATGTT....................................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................AGTCCAAGATAAAGTGAGGG............................................................ | 20 | 3 | 6 | 0.33 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................TCCGTATTAGGTTGCCTATC......... | 20 | 3 | 6 | 0.33 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| .....................................................................................................................................................................................................................AGTGATAAAAGTAACGAGAGAAA........................................................................................................................ | 23 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................CCTTACGGGGGTCGGGAGT........................................................................................................................................................................................................................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................................................................................................AAGTTCATGATAAAGTGAGGG............................................................ | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................GGGAAGGTATGTCTTGG.............................................. | 17 | 2 | 12 | 0.17 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................TCCGTATTAGGTTGTG............. | 16 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................GTGATAAAAGTAACGAGAGAAA........................................................................................................................ | 22 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................AAGTCCAAGATAAAGTGAGG............................................................. | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................CTTGTTAGGTTGTGAATC......... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................TCTTTGAGATGTCCTGG.......................................................................................................................................................................................................................................... | 17 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................TTCCGTATTAGGTTG............... | 15 | 1 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................AGTCCAAGATAAAGTGAGG............................................................. | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................CCGTGTTAGGTTGCGAA........... | 17 | 2 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................ACGATCTTTGAGATG................................................................................................................................................................................................................................................ | 15 | 1 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................TTAAGTCCAAGATAA.................................................................... | 15 | 1 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................GGGAATTCAAAGTCGAAA............................................................................................................................................................................................................................................................................... | 18 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................CTTGTTAGGTTGTGT............ | 15 | 1 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................AGGATCTTCCTTGTTGGGT................. | 19 | 3 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................AGATAAAATACGGGC........................................................... | 15 | 1 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................GCAACAAGGTATATAGATG......................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................................................................................................................................................................................................................................................CTCCGTGTTAGGTTGC.............. | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................TGGTCTAGTGATACA...................................................................................................................................... | 15 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................AGTTCTAGATAAAGTGAGG............................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3r:16381665-16382020 + | dsi_13996 | TGCAGTGCGTCCAGCAACTGGATCTGGATGCCATAAAGC---CAGGCG-ATCAGGTGTG----------TAATGCCC------GC-AGCCCTCAAGTTTCG----------GCTTTTGTTCGGTTTA-CATCT-----------TA-------CTAGAAACTCT------------------------------------------------------------------------------------------------------------------------------------------------------AC---AGCACAGCTAGCACATTGATGTTTGTCAGTTATTCGT---GGAAAAACTTACA--TAC------GTTGTTGCAAATATA----TACAATC--C-------------ATA----CATTTATTGAT-TCCCAGATCACTATTTTCATTACCCTCGTT----TAAA-----------CATT-GTT------ATTATTTTGAGCAAAGA--CATC-----------------------------------------------------------------------------GC-TAAAATTCAAGTTCT-A-TTTTA-TTCCC--GTAC--ATACAGAACCGGACGGAAACTCCCAGAAGGCACAATCCAACACATAGTCACA------------GCCA |
| droSec2 | scaffold_0:17247020-17247375 + | TGCAGTGCGTCCAGCAACTGGATCTGGATGCCATGATGC---CAGGCG-ATCAGGTGTG----------TAATGCTC------GC-AGCCCTCAAGTTTCG----------GCTTTTGTTCGGTTTA-CATCT-----------TA-------CTTGAAACTCT------------------------------------------------------------------------------------------------------------------------------------------------------AC---AGCACAGCTAGCACATTGATGTTTGTCAGTTATTCGT---GGAAAAACTTACA--TAC------GTTGTTGCAAATATA----TACAATC--C-------------ATA----CATTTATTGAT-TCCCAGCTCACTATTTTCATTACCCTTGTT----CAAA-----------CATT-GTT------ATTATTTTGAGCAAAGA--AATC-----------------------------------------------------------------------------GC-TAAAATTTAAGTTCT-A-TTTTA-TTCCC--GTAC--ATACAGAACCGGACGGAAACTCCCAGAAGGCACAATCCAACCCATAGTCACA------------GCCA | |
| dm3 | chr3R:4609833-4610211 - | TGCAGTGCGTCCAGCAACTGGATCTGGATACCATGATGC---CAGGCG-ATCAGGTGTG----------TAATGCCC------GC-AGCCCTCAAGTTTCG----------GCTTTTGTTAGGTTTA-CACCTTACTAGCAACTTA-------CTAGAAACTCC------------------------------------------------------------------------------------------------------------------------------------------------------AC---AGCACAGCTAGCACATTAATGATTGTCAGATATTTGT---GGAGTAACTTACA--TAC------GTTGTTGCAAATATA----TACAATC--C-------------ATA----AATTTATTGAT-TCCCAGCTCACTATTTTCATTACTCTTATT----TAAA-----------CATT-GTT------AATATTTTGAGCAAAGA--AATC-----------------------------------------------------------------------------GC-TAAAATTCAAGTTTT-A-TTTTG-TTCCC--GTAC--ATACAGAACCGGACGGAAACTCCCAAAAGGCACAATTCAACCCACAGTCACAGTCACAGTCACAGCCA | |
| droEre2 | scaffold_4770:16976393-16976757 + | TGCAGTGCGTCCAGCAGCTGGATCTGGACGCCATGATGC---CAGGCG-ATCAGGTGTG----------TAACGCCC---------------------TTG----------GTCCCTGTTATGTTGA-AATCT-----------TTTCCGTATTCACAAACTCC------------------------------------------------------------------------------------------------------------------------------------------------------AC---AGCATAGCTAGCATATAAATGTTTGTCAGTTATTCGT---GGAAAGGCCTACA--GACTTTCATGTTGTTGCAAACATA----TACAACC--C-------------GTA----TATTCATTGAG-TCCCAACTCACTATTTGCATTAACCTTGCTTGTTTAAAAAC----CA-CATAT-TTT------CTATTTTTGGCCAACGA--GATC-----------------------------------------------------------------------------AC-TAAAATTCCAGTCTT-A-TTTTA-CTCCC--GTCC--ACACAGAACCGCACGGAAACTCCCAGAAGACACAATCCAGCACATAGTCACA------------GCCA | |
| droYak3 | 3R:8671186-8671498 - | TGCAGTGCGTCCAGCAACTGGATCTGGATGCCATGATGC---CAGGCG-ATCAGGTGTG----------TAACGCCC------------------------CTTA------GCCCCAA----------------------------TACATATTCAGAAACTCC------------------------------------------------------------------------------------------------------------------------------------------------------AC---AGCATAGCTAGCACATTAATGTTTGTCAGTTATTCGT---GGAAAACCTTACA--GAC------GTTGTTGT--------------------C-------------ATA----TATTTATTGAG-TCCCAACTCACTATTTGCATTACCCTTACTTGTTTACA-----------------------------ACGAGGACAACGA--GGTC-----------------------------------------------------------------------------GC-TAAAATTCAAGTCTT-A-TTTTC-TTCCT--ATCC--ACACAGAACCGGACGGAAACTCCCAGAAGGCACAATCCAGCGCATAGTCACA------------GCCA | |
| droEug1 | scf7180000409490:112777-113099 + | TGCAGTGCGTCCAGCAATTGGATCTGGATGCCATGATGC---CAGGCG-ATCAGGTGAG----------AAAAGATA------G--------------------------------------ATTGA-GTT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT---ATCCCAGATGGCTCATTTATGTTTGTCAGTTAACCGT---AGAAA-CTTTATA--AAG------AGTTTGGCAAACAGAACA-AAAAAAC--A-------------ATA----TATTTATTGAT-AGCCAACT---TATTTTCATTATCTTTATTTGTTTTAAC------CAACACTT-ATG------AAAGTTTCGGACTAAGG--AGTT-----------------------------------------------------------------------------CC-TAAAATTCAAGTTTTTG-TTTCG-TTTTC--CATC--ACACAGAACCGGACGGAAGCTACACGGAGGCACAACCCAACCCAAAGTCACA------------ATCA | |
| droBia1 | scf7180000302402:7321131-7321382 - | AGC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTAGCTAGCCCATTTATGTATGTCAGTTAACCGT---AGACAGCTTTACT--GAG------TGTTTTGCAAACATACAAATACAATC--C-------------ATA----TATTTATTGAT-ACCCCAGT---TATTTCCATTAGCTTCATTCGTTTTAGC------CA-CCCTT-ATG------AAAATTTCGGACTAAGA--AATC-----------------------------------------------------------------------------GC-AAAAATTCAAGTTTT-G-TTTTA-TTTTTC-GCCCAAAAACAGAACCGCACGGAAACCCACCGGAGGCACAACCCAACCCAAAGTCATA------------GTCA | |
| droTak1 | scf7180000414009:214606-214855 + | ACTA--------------------------------------------------------------------------------------------------------------------------------------------------------TAAATCCC------------------------------------------------------------------------------------------------------------------------------------------------ATCATTTT---AGCATAGCTAGCCCATTTATGTTTGTCAGTTACTCTT---AAAAAACA-AAAGCAGAG------AGTTTTGCGAGCATA----TACAACC--C-------------ACA----TATTTATTGAT-AGCCAACA---TATTTCCCTTAGCTTTACTCGTTTTAAC------CG-CCCTT-ATG------AATATTTTGGACTAAGA--AATC-----------------------------------------------------------------------------GC-TAAAATTTAAGTTTC-G-TTTTA-TTTTTCCACGC--ACACAGAGCCACCAG---------CGAAGGCCCAA---------CAACCCAA------------CCCA | |
| droEle1 | scf7180000491212:1188109-1188526 + | TGCAGTGCGTCCAGCAACTGGACCTGGATGCCAAGATGC---CAGGCG-ATCAGGTGTG----------TATAAGCC------------------------CTT--------------------------------------------------------------------------------------------------CTAGCATTGAGCTAATTTTACTTGACGTTTAAAACTGAATTGTTAACCCGAATATAACTTTTGTCTGAAAGTCACCTCATTCTCCCCATCTCTAAATATAAATCATATTTCATT---ATCATGGCTACCTCAGAAATGTCTGTCAGTTAATCGT---AGAAAACACAACG--TAG----------------ACATA----TACAATC--T-------------ATATACAAAGATACTGAT-ACCAAACT---TCT-TCCTTTTTCTTTAGTTGTTTTAAC------CA-GCTTA-ATG------CAAATTTCGGACTAAGAAAAATC-----------------------------------------------------------------------------GC-TTAAATTAAAGTTTT-G-TTTTA-TTTGTT-TGCT--ATACAGAACCGGACGGAAACTCCGCGGAGGCACAACCCAACTCAAAGTCACA------------A--- | |
| droFic1 | scf7180000454106:2558885-2559216 + | TGCAGTGCGTCCAGCAACTGGACCTGGACGCCATGACGT---CAGACA-ATCAGGTGGG----------TGAAACTCCA-GTTGAAGTA------------------------------------------CT-----------TA-------ACCCAACTTCA------------------------------------------------------------------------------------------------------------------------------------------------------AT---ACCACAGCCATCCCAGTCATGTTTGTCAGTTGATTGTAGTGGAAAACGTAACG--GAG------TGTCTTGCCAACATA----TACAACC--C-------------ACA----AAGAT-----T-ACCCAACT---TGT-TCCCTTAC-TTTAC-CTTTTTAAC------CT-ACTTG-ATG------AACCTTTCGGACTAAGG--AATC-----------------------------------------------------------------------------CC-TAAAATTCACGTTGT-G-TTGTA-CTCTTT-CCCC--ACACAGAACCGCACGGAGACTCCCCGGAGGCACAACCAAACCCAAAGTCACA------------ACCA | |
| droKik1 | scf7180000302247:283475-283891 + | TCATATAAATTTACAAGCTATGTTTTGATTCTATTCCTT---CTGATCCATAGTCTGTT----------TCAGTTACTT-GTTGAAGATCTTTAAATCTTA----------AATTTAATTAAATTTGGTAGTTTGTTAGGATATTT-------TTGTGAAATAT-----------------------------------------------------------------------------------------------------------------------------------------AAAAGATACTTTCTTATTTTCTGAGCCACCCCAGTCCAGTTTGTCAGTTATCTGT---AGAAAGTCATTCGTAGAA------TGTGTTACGGACACACACACACACCCTT-CTTTATTT-----AGATATATAAACACGGAT-ATCCCAGT---TATTCTCA-CACTTTTGCTTGTTCAAAT------GA-CCTTT-TTG------AT-----CAGATATCGC--GACCGCGTTTTCCGC------------------------------------------------------------------GCAAAAAATTAAAGTTTT-T-TACTA-TTTT---TCGC--ACCCAGAAC---ACGCACGCCCGACGGCCGAGTAACCAAACCCAAAGTCACA------------GCCA | |
| droAna3 | scaffold_13340:11019733-11020075 - | TGAACTGCGTCCAGCAGCTCGACCTCGATTCGATGATGA---CAGGAG-AACAGGTGCG----------TAAAATTTACTAT-------------------------------------------------------------------------TGAAACCCA------------------------------------------------------------------------------------------------------------------------------------------------------AGACAGGCGTAGGTAGCCCAGTCAAGTTTGTCAGTTGATCGA---AGAGCCCTAAACG--TAG------AATTAAGCGAATAAAAACTTACAATATT-CAATATTTATTTT-GATA------TATTGGT-AGTCGAGC---CACATCCA-CAGATTTTCTTATTTTAAC------CA-AACTT-TCA------AATTTTTCGATTATCGC--GATCGGTACTTCCCG------------------------------------------------------------------GC-TAAAATTCAAGTTTT-G-TTTTA-TTTTC--CCGC--ACATAGAACCACACGGAATCCCCACGAAGGCACC---------ACAATCAGA------------ACCA | |
| droBip1 | scf7180000396374:62934-63264 + | TGCACTGCGTTCAGCAGCTGGACCTGGATTCAATGATGA---CAGGAG-AACAGGTGGG----------TAAAATAT-------------------------------------------------------------------AA-------CTATATGTAGA------------------------------------------------------------------------------------------------------------------------------------------------------CA---AGCCTAGGTAGCTCAGTCAAGTTTGTCAGTTGATCAG---AGAACCCTAGACG--TAG------AATTATGCGAATAAGAACTCACAATATT-ATATATTTATTTA-GATA------TATTGGT-AGTCGAGC---CACTTCCA-CAGATTTTCTTATTTTAAC------CA-CACTTTCCC------AACCTTTTGATTATCGC--GATCGGTATTGTCCG------------------------------------------------------------------GC-TAAAATTCAAGTTTC-G-TTTTA-TTCCC--GTCC----GTAGAACCCCACGGAGAGCCCACGAAGGCACCAAC----------------------------ACA | |
| dp5 | 2:27265934-27265993 - | TGCAGTGCGTCCAGCAATTGGATCTGGATGCCATGATGCCGTCAGCCG-AACAGGTGTG----------TA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_6:2555961-2556020 - | TGCAGTGCGTCCAGCAGTTGGATCTGGATGCCATGATGCCGTCAGCCG-AACAGGTGTG----------TA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004902:4607456-4607760 - | TTCAGTGTGTGCAACAACTCGATCTTGACGCCGGGATGC---CTGTTG-ATCAGGTGAG----------TTTCAGTT------------------------TTAAATGTCATATACAAAAAAA------------------------TAATATTAAAAAACACCAAAACAAAAAAAAAACACATTTATTTCCATCAACATTTATGGCCTTACATGGCTTTTAATACACATTTTAAATAAACTTGCTACCCCT--------------------------------------------------------------------------CAACCCCAAACATTTTTG-----------------------------------------------------------------------------------------------------------------------AAACAATTGATTGTATAAAATTTAACCA-AAGTG-AAATATTTTGTCT-------------------------------------------------------------------------------------------------------------TTTTC-GTTTTTTGTTTTC--T-GC-AGAGCAAATCCGAGCGT--------ACAAGGCACAATC------------------------------- | |
| droVir3 | scaffold_13047:1869996-1870029 + | TGATGTGCGTACAGCAACTGGATCTGGATGCGAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droMoj3 | scaffold_6540:3800419-3800763 + | TGCTGTGCGTGCAGCAGCTCGATCTAGATGCGGTCGTGGAG-CAGGAG-AGCTGGCAGTCAACGGTGCGTATATACT------------------------CTAAACGAAATGTACAA----ATTGA-GTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TG---AGTGTAGAAGGTGTGTCTGTCTGTGTCGGT---------------------------------------------------------------GTGTGTGTGTTTA-GATG------TAGTCAGAAGTCAGTC---AGTAGTTA-GGAATTTTTTGATTTTG----------------------------------AGTGATCGA--GATTTATATTTTGACCCCAGCAGAGGCAGACAGTTTTTTAATCCTTAAGAAAATCAATAACCACACATAAACATTTCTAGAAC-ATAAATTCAAGTGCG-G-CTTCT-TGCTC--TTGC--AGACGAA----GGCGGAACCACGGACGAGGCACAGTC------------------------------- | |
| droGri2 | scaffold_14906:8705943-8705976 + | TGCTGTGCGTGCAACAGCTCGATCTGGATGCGAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/19/2015 at 01:07 AM