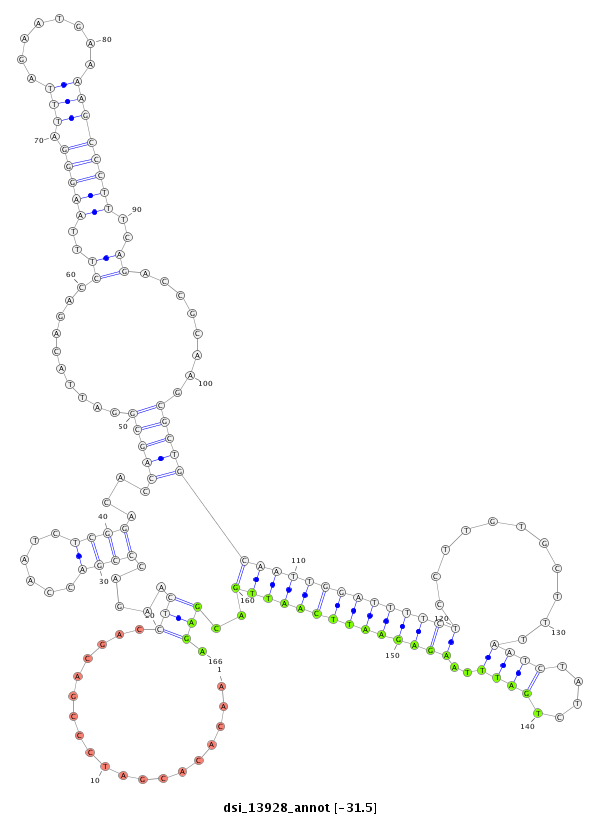
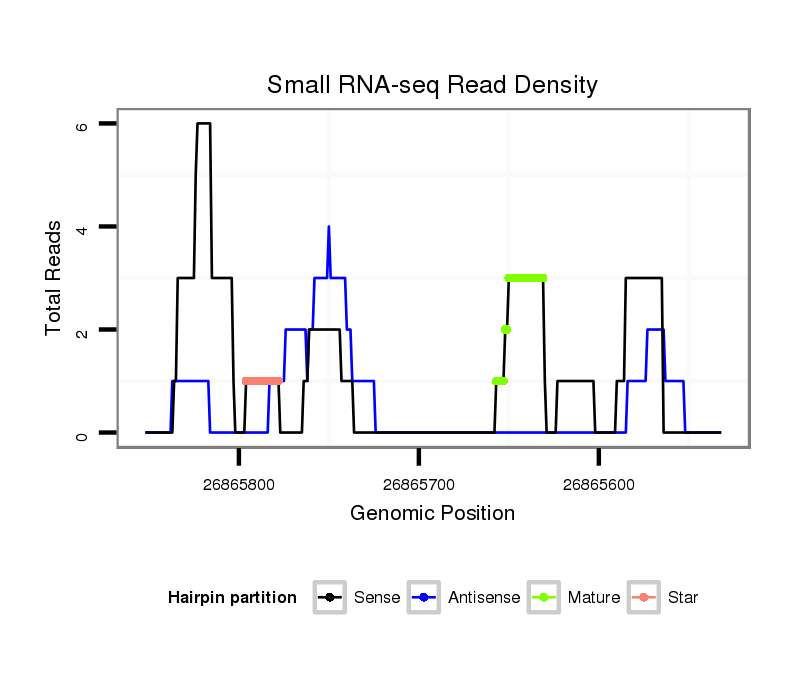

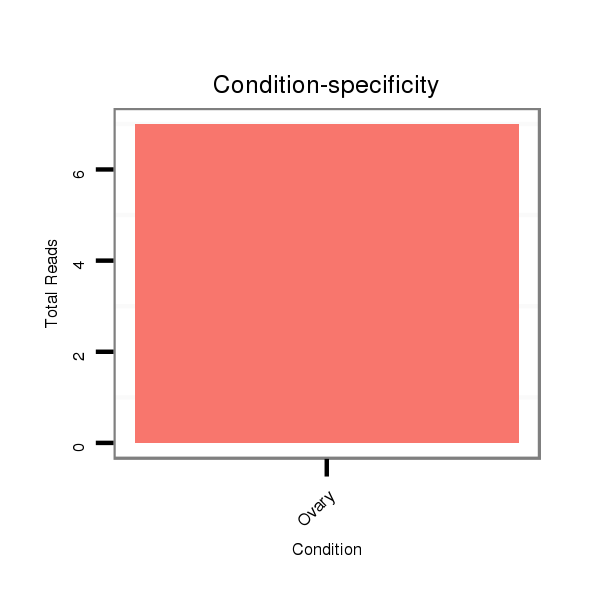
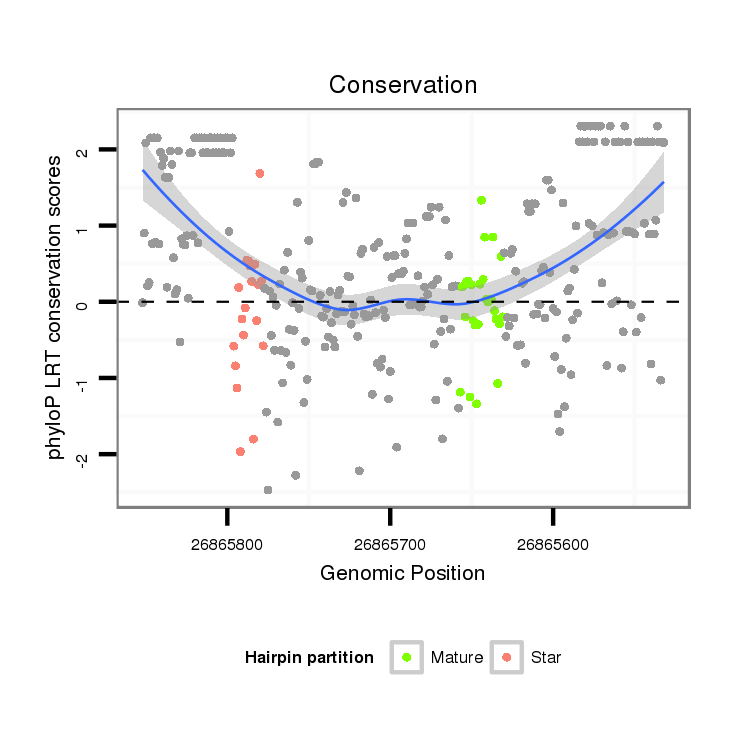
ID:dsi_13928 |
Coordinate:3r:26865582-26865802 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -31.4 | -31.3 | -31.2 |
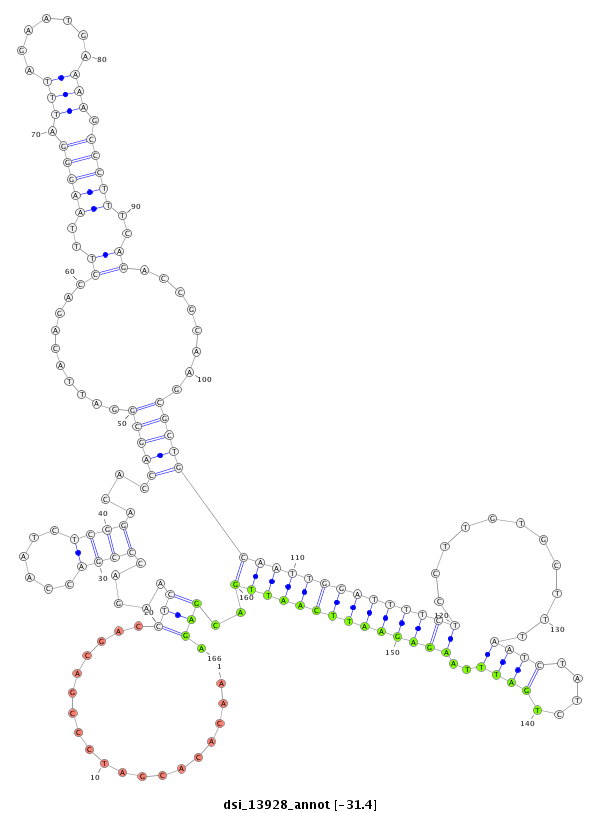 |
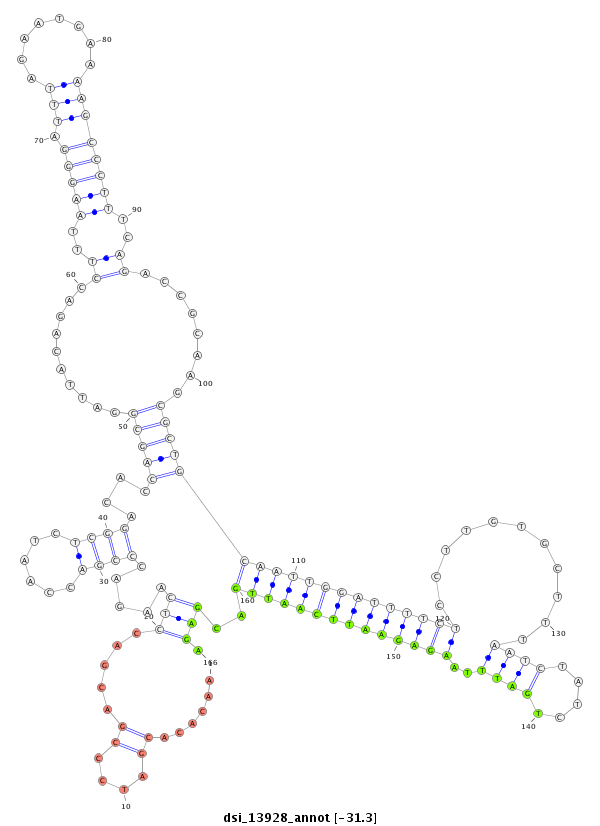 |
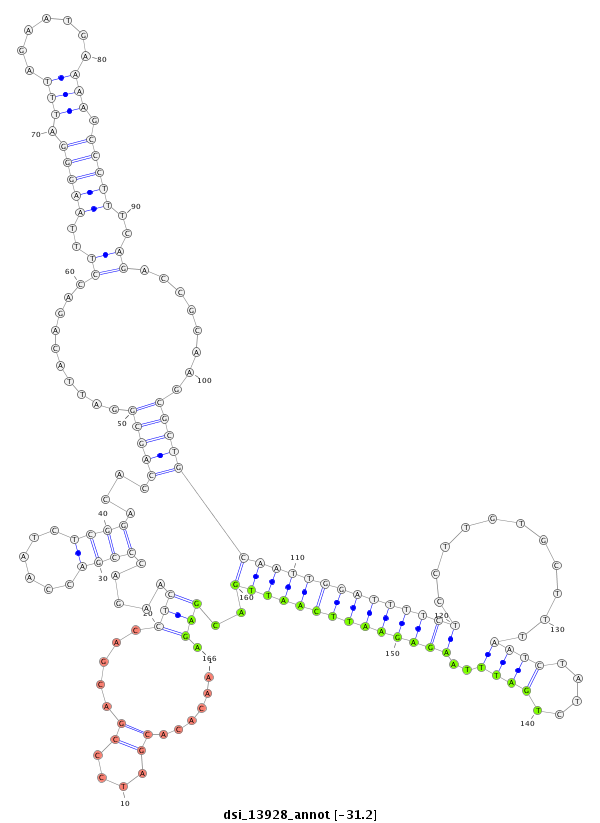 |
CDS [3r_26865803_26865820_-]; five_prime_UTR [3r_26865821_26865921_-]; exon [3r_26865803_26865921_-]; CDS [3r_26865042_26865581_-]; exon [3r_26864547_26865581_-]; intron [3r_26865582_26865802_-]
No Repeatable elements found
| ##################################################-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CCCCAACAAACACACTCACCCGAATCGTCGCTATGAACTCCTTCTCAGTGGTAAGTAACACACGATCCCGACGACCTCAAGACCCGACCAATCTCGGACACCAGCGGATTACAGACCTTTAAGGGATTTAGAATGAAAAGCCCTTTCAGACCGCAAGCGCTGCAATTGGATTTTCTCCTTGTGCTTTAATCTATCTGATTTAAGAGAATTCAATTGACGAGATCACCAGTTTGATATGGAATTTGCACTAACCTCGACTACCTACTTTCAGTTCTTAGTGTTCGCGCTGGCCGCTCTGGCTGCAGCAGAGCCACCATCCGG ********************************************************...................(((.....((((......))))....(((((..........((..(((((.(((........))))))))..))........)))))((((((((((((((...........((((.....))))..))))))))))))))..))).*************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
M025 embryo |
SRR618934 dsim w501 ovaries |
M024 male body |
M023 head |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................CGCTATGAACTCCTTCTCAGT................................................................................................................................................................................................................................................................................ | 21 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................CCCGAATCGTCGCTATGAA............................................................................................................................................................................................................................................................................................ | 19 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................TCAGTTCTTAGTGTTCGCGCT................................. | 21 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................TTCAATTGACGAGATCATCAGTTTGATA..................................................................................... | 28 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TGACGAGATCATCAGTTTGATATGGAA................................................................................ | 27 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................CACCCGAATCGTCGCTATGAA............................................................................................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................TGATTTAAGAGAATTCAATTGACGAGA................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................AACACACGATCCCGACGAC...................................................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................ATGACCTCCTTCTCAGTGGT............................................................................................................................................................................................................................................................................. | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CAATCTCGGACACCAGCGGAT.................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................AGAGAATTCAATTGACGAGAT.................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................TAAGAGAATTCAATTGACGAGA................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................TACTTTCAGTTCTTAGTGTTCGCGCT................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................GCTATGAACTCCTTCTCAGTG............................................................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TCTCGGACACCAGCGGATTACAGAC............................................................................................................................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................TTTGATATGGAATTTGCACTA....................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................CTATGAACTCCTGCACAGTGGCA............................................................................................................................................................................................................................................................................ | 23 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................TATGGAACCTGCACTAGCCT................................................................... | 20 | 3 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ......................................TCCTTCTGAGTGGTTAGTA........................................................................................................................................................................................................................................................................ | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................ATGAACTCCTGCACAGTGGCA............................................................................................................................................................................................................................................................................ | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................AGGAGATCACGAGTTTGA....................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................AGTGGTAAGTAACAGACA................................................................................................................................................................................................................................................................. | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................................ATCGGTTTTAAGAGAATT............................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................................................................................CGATTGTCGCTCTGGCTGCA................. | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................................................................................ATTTTCTCCTCGTGCT........................................................................................................................................ | 16 | 1 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................ACTGCATTTGGATTTTCTC................................................................................................................................................ | 19 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................CCTCTCTCGGACACCA.......................................................................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................TAGGATTTACTCCTTGAGC......................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................ATCACCAGGTTAATAGGGA................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GGGGTTGTTTGTGTGAGTGGGCTTAGCAGCGATACTTGAGGAAGAGTCACCATTCATTGTGTGCTAGGGCTGCTGGAGTTCTGGGCTGGTTAGAGCCTGTGGTCGCCTAATGTCTGGAAATTCCCTAAATCTTACTTTTCGGGAAAGTCTGGCGTTCGCGACGTTAACCTAAAAGAGGAACACGAAATTAGATAGACTAAATTCTCTTAAGTTAACTGCTCTAGTGGTCAAACTATACCTTAAACGTGATTGGAGCTGATGGATGAAAGTCAAGAATCACAAGCGCGACCGGCGAGACCGACGTCGTCTCGGTGGTAGGCC
***************************************************************************************************...................(((.....((((......))))....(((((..........((..(((((.(((........))))))))..))........)))))((((((((((((((...........((((.....))))..))))))))))))))..))).******************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M053 female body |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553485 Chicharo_3 day-old ovaries |
M023 head |
SRR553486 Makindu_3 day-old ovaries |
M025 embryo |
M024 male body |
SRR1275487 Male larvae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................CCTGGAGTTATGGGCTGGTAA..................................................................................................................................................................................................................................... | 21 | 3 | 1 | 8.00 | 8 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................TCCTGGAGTTATGGGCTGGTAA..................................................................................................................................................................................................................................... | 22 | 3 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................CGTCACCATTCATTGTCT................................................................................................................................................................................................................................................................... | 18 | 2 | 5 | 2.20 | 11 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 2 | 0 | 0 |
| ..................................................................................................GTGGTCGCCTAATGTCTGGAG.......................................................................................................................................................................................................... | 21 | 1 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................CGCCTAATGTCTGGAGATTCC..................................................................................................................................................................................................... | 21 | 1 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................CTGGAGTTATGGGCTGGTAA..................................................................................................................................................................................................................................... | 20 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................ATTCCCTAAATCTAAC.......................................................................................................................................................................................... | 16 | 1 | 7 | 1.71 | 12 | 0 | 3 | 2 | 0 | 0 | 0 | 7 | 0 | 0 | 0 |
| ...................................................................................................................AGCAATTCCCTAAATCTAAC.......................................................................................................................................................................................... | 20 | 3 | 5 | 1.60 | 8 | 0 | 4 | 2 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| ........................AGCGGCGATACTTGCG......................................................................................................................................................................................................................................................................................... | 16 | 2 | 20 | 1.45 | 29 | 0 | 0 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................GTCAAGAATCACAAGCGCGAC................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................GGTCGCCTAATGTCTGGAGAT........................................................................................................................................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................CTGCTGGAGTTCTGGGCTGGT....................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................GCCTGTGGTCGCCTAATGTCT.............................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............AGTGGGCTTAGCAGCGATACT............................................................................................................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................CCTGGAGTGATGGGCTGGTTA..................................................................................................................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................TTCTGGGCTGGTTAGAGCCTGTGGT.......................................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................CTGGAGTGATGGGCTGGTAA..................................................................................................................................................................................................................................... | 20 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................CAAGCGCGACCGGCGAGACCG..................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................CTTGGCAGCGATACTTG........................................................................................................................................................................................................................................................................................... | 17 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................TCGCCTAATGTCTGGAAATTCCCTAA................................................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................AGAGCCTGTGGTCGCCTAATG................................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................ACGTGATTGGAGGTGATGGAT......................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................GCAATTCCCTAAATCTAAC.......................................................................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................TCCGGGAGTTATGGGCTGG........................................................................................................................................................................................................................................ | 19 | 3 | 8 | 0.75 | 6 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | 1 |
| ........................................................................CGGGAGTTATGGGCTGGTAA..................................................................................................................................................................................................................................... | 20 | 3 | 6 | 0.67 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................GCAATTCCCTAAATCTAACAT........................................................................................................................................................................................ | 21 | 3 | 3 | 0.67 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................AATTCCCTAAATCTAAC.......................................................................................................................................................................................... | 17 | 1 | 5 | 0.60 | 3 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................TCCTGGAGTGATGGGCTGG........................................................................................................................................................................................................................................ | 19 | 3 | 8 | 0.38 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................CTGGAGTGATGGGCTGGG....................................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................CGGGAGTGATGGGCTGGTTA..................................................................................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................AATTCCCTAAATCTAACAT........................................................................................................................................................................................ | 19 | 2 | 6 | 0.33 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................GCAATTCCCTAAATCT............................................................................................................................................................................................. | 16 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................CGGGAGTTATGGGCTGGG....................................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.30 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 |
| .........................GCAGCGATACTTGCGGA....................................................................................................................................................................................................................................................................................... | 17 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................TCGCAGCGATACTTG........................................................................................................................................................................................................................................................................................... | 15 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................TTCCTGGAGTGCTGGGCTGG........................................................................................................................................................................................................................................ | 20 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................GTTCCTGGAGTTATGGGCTG......................................................................................................................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................TTGCCGCTGATGGATGAGAG.................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................TTGCAGCGATACTTG........................................................................................................................................................................................................................................................................................... | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................TGAGAGGAGCTGATGGAT......................................................... | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................GCAGCGATACTTGTG......................................................................................................................................................................................................................................................................................... | 15 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................GTTATGGGCTGGTTA..................................................................................................................................................................................................................................... | 15 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................TTGCCGCTGATGGATGA....................................................... | 17 | 2 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................GCAGCTATACTTGAG......................................................................................................................................................................................................................................................................................... | 15 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................ATTCCCTAAATCTAACAT........................................................................................................................................................................................ | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................GCTCTGCAGCGATACTTG........................................................................................................................................................................................................................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................GCAGCGATACTTGCGGT....................................................................................................................................................................................................................................................................................... | 17 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................AGCAATTCCCTAAATCTAA........................................................................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................CTTGCAGCGATACTTG........................................................................................................................................................................................................................................................................................... | 16 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................TTGCCGCTGATGGATGAGA..................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................AGTCCGGCGCTCGCGACGC............................................................................................................................................................. | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................ATTCCCTAAATCTAA........................................................................................................................................................................................... | 15 | 1 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................AATTCCCTAAATCTA............................................................................................................................................................................................ | 15 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................GCGCTAGTGGTCAAA......................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........TGTGTGAGTGGTCTT......................................................................................................................................................................................................................................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................TTAAGGTGATTGGAGC................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................GGGAGTTATGGGCTGGTAA..................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3r:26865532-26865852 - | dsi_13928 | CCCCAACA-------AACACACTCACCCGAATCGTCGCTATGAACTC-CTTCTCAGTGG-TAAGTAACAC-ACGATCCCGACGACCTCAAGA-CC------------------------------CGAC---------CAATCTCGGACACCAGCGGATTACAGACCTTTAAGGGATTTAGAATGAAAAGCCCTTTCAGACCGCAAGCGCTGCAATTGGATT--------------T------------------------------------------------TCTCCTTGTGCTTT---AA-TCTATCTGAT------------------------------------------------------------------------------TTAAGAGAATTCAATTGACGAGA-TCACCAGTTTG-------ATATGGAATTTGCACTAACCTCGACTA-CC---TACTTTCAGTTCTTAGTGTTCGCGCTG--GCCGCTCT-------------GGCTGCAGCAGAGCCAC---CATCCGG |
| droSec2 | scaffold_31:244534-244868 - | CCCCAACA-------AACACACTCACCCGAATCGTCGCTATGAACTC-CTTCTCAGTGG-TAAGTAACAC-ACGATCCCGACGACCTCAAGA-CTCCAGAC---------CC-----CA--CGGCCGAC---------CAATCTCGGACACCAGCGGATTACAGACCTTTAAGGGATTTAGAATGAAAAGCCCTTTCAGACCGCAAGCGCTGCAATTGGATT--------------T------------------------------------------------TCTCCTTGTGCTTT---AA-TCTATCTGAT------------------------------------------------------------------------------TTAAGAGAATTCAATTGACGAGA-TCACCAGTTTG-------ATATGGAATTTGCACTAACCTCCACTA-CC---TACTTTCAGTTCTTAGTGTTCGCGCTG--GCCGCTCT-------------GGCTGCAGCAGAGCCAC---CATCCGG | |
| dm3 | chr3R:27590021-27590419 - | CCCCAACA-------AACACACTCACCCGAAACGTCGCTATGAACTC-CTTCTCAGTGG-TAAGTAACAC-ACGATCCCGACGACCTCAAGA-CCCCAGAC---------CC-----CA--GAGCCGAT---------TAAACCTGGACGCAAGCGGATTACACCCCTT-ACGGGATTTAGAAAGAATAGCCCTGTCAGACCGCAAACGCTGCAATTGGATT--------------T-CAAAA-AAACTTCAA-------------------TAGTTTCAAAAACTCTCCTTGTGCCTT---AA-TCTATGGGAT--------------------------------------TTCAATTCACATTCCACAAAAAGAC-GCATT-CGAGTTATTTAAGAGAATTCAATTGATGAGA-TCACCAGTTTG-------ATATGGAATTTGCACTAACCTCCACTA-CC---TACTTTCAGTTCTTAGTGTTCGCGCTG--GCCGCTCT-------------TGCTGCAGCAGAGCCAC---CATCCGG | |
| droEre2 | scaffold_4820:380329-380565 + | CCCCAACA-------AACACACTCACCCGAATCGTCGCTATGAACTC-CTTCTCAGTGG-TAAGTAACAC-------CAGGCGA----------------------------------------CCG---------------TTGGGACTCAAACGGATTACAGACCTGAAAGGGATTGAGAGGGAATAACT---------------CGCTGCAATTGGATT--------------T-AAAA-------------------------------------------------------------------------------------------------------------------------------------------------------------G---------------------CACTGGA-------ATATGGAATGTGCACTAACCTCCAGTA-TC---TACTTTCAGTTCTTAGTGTTCGCGCTG--GCCGCGCT-------------GGCTGCAGCAGAGCCAC---CATCCGG | |
| droYak3 | 3R:28524494-28524854 - | CCCCAACA-------AACACACTCACCCGAATCGTCGCTATGAACTC-CTTCTCAGTGG-TAAGTACCAT-ACGATCCCGACGATCAG------------------------------------CCATCCTCCA--AA----TTGGGACTCAAACGGATTACGGACCTTAAAGGAATTTAGAAAGAGTTAATCTTTCGGACGCCAATCGCTGCAATTGGATT--------------T-GAAAT-TAACTGAAA--------------------AGGTTCAAAGACTCTCCTTGTGCCTT---CA---------------------------------------------------------------------AAGAT-GCAGTACCAGTTCTTTGCCAATATTCAATTGACAAGA-TCACCAGTTTGGTGGCCCATATGGAATGTGCACTAACCTCCACTT-TC---TCCTTTCAGTTCTTAGTGTTCGCGCTG--GCCGCTCT-------------GGCTGCAGCAGAGCCAC---CATCCGG | |
| droEug1 | scf7180000409488:313382-313718 + | CCCCAACA-------AACACACTCACCCGACTCGTCGCTATGAACTC-CTTCTCAGTGG-TAAGTAGCCT-ACGATCCTGAAGTCCAAA-GA-CCT--G-----------------------A----TCTTGCA--AA-----TCGGTCTTAAACGGATAAAAGACTTTAAGCGGAATTAAAA------GCT------------AA-------------ACATCTTCATTTGGACCTAGAAAC-TAG------------------------------------------CTTTTGTTTTCTTTAAA--------CATAAAAGCAACTTTTTATAACAAATAGCAGA--AGGAT-------------------------------CGCTTTTCTTTCCCGAACTTCAGTCGA---------------------CCAACTTGGACTATCCACTAACCTCTCGTT-C-----CCTTTCAGTTCTTAGTGTTCGCTCTG--GCTGCTTT-------------GGTCGCGGCAGAGCCAC---CATCTGG | |
| droBia1 | scf7180000302035:255337-255655 - | CCCCAACA-------AACAC-----------TCGTCGCTATGAACTC-CTTCTCAGTGG-TAAGTGGCAC-TCGATCTTGGAGGTTGAA----CTCC---------------------------------------------TCGGGACCTTTACGGATATCGGACTGTAAACGGATTTAAAT-----AGCACCTTGGG------------------------------------------AC-CAACCGAAAAAGAAGATCAATTCTCTATTAG-TTCAAAAA-TCTCCTCCTGC------------------TACAAAAGCAAGCTTCTAT------TGGGAGACTAGGAC-----------------------------------CTACTCCGCTGATCTTCAGCTG----------------------CCACTGTGGACTA-CCATTAACCGCGTGTT-C-----CTTTGCAGTTCTTAGTGTTCGCTCTG--GCCGCTCT-------------GGCTGCGGCTGAGCCAC---CGTCCGG | |
| droTak1 | scf7180000415712:680364-680479 - | CCCCAACA-------AACACA---------ATCGTCGCTATGAACTC-CTTCTCAGTGG-TAAGTGA--------------AGATCTGAAGA-TTCGAAAA---------------------------CCAACC--CA-----TCGAACCTAAACGGATAACGGACTATAAACGGATTTAAAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEle1 | scf7180000491194:1641018-1641155 - | TCCCAACA-------AACACACTCACCCAACTCGTCGCTATGAACTC-CTTCTCAGTGG-TAAGTAATAA-ACGATTCCGAAGATCTCAAGG-CCCA-GG---------------------CAGCCGACAGAAAAAGA-----GTGCACCTAAACGGATAACGGATTTAAAACGGA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000779322:99478-99621 + | CCCCAACA-------AACACACTCACCCAACTCGTCGCTATGAACTC-CTTCTCAGTGG-TAAGTGATAG-AGGATCCTGAAGATCGCAGGA-CTCAAGG---------------------CAGCCGACCAAAA--CA-----TTGGACCTAAACGGATAACGGGCCTAAAACGGATTTTAAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000454097:15123-15511 - | CCCCAACA-------AACACACACACCCGACTCGTCGCTATGAACTC-CTTCTCAGTGG-TAAGTGAACT-TCTGTCCCGGAGACCTCCGAC-CTCCTTCC---------C------CAGTCATCCAGC----AACCA-----TTGGACCCGAACGGATAGCGGACCCCAAACGGATAAA-AGGGAAT--CCCCATAAATCA---------G-AGATAACTT--------------C-CAAACTAAATTCCAC-------------------TAAACACGAAATCTCCCCAAATGGGA---------------------------------------------AGACACAGATTTCAAAATAATTTCTTCAAAAAGGCTTCTCAATCACTGCCTTGCCAGACTTCGGTTGCCTCGGATGTACAG--------CCAATGTGGACTACCTATTAACGAGCCGTGTTG---TCTTTGCAGTTCTTAGTGTTCGCCCTG--GCCGCCTT-------------GGCTGCGGCGGAGCCTC---CATCCGG | |
| droKik1 | scf7180000301641:73538-73685 - | CCCCAAAACTC----AACACACACACCCAAATCGTCGCTATGAACTC-CTTCTCAGTGG-TAAGTAGGATCAGGATCCAGAA-ATCTACAGATCACAAAAA---------ACGGATA-------ACTGC---------CACTTTTGGATAACAGAGG---GAAAGCCTTGGAAGCATTAGGAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_12911:3494681-3494738 + | A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTACAGTTCTTAGTGTTCGCTCTG--GCCGCCTT-------------GGCTGCGGCTGAGCCGC---CATCTGG | |
| droBip1 | scf7180000396640:358192-358250 + | T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTTGCAGTTCTTAGTGTTCGCTCTG--GCCGCTTT-------------GGCTGCGGCTGAGCCGC---CATCTGG | |
| dp5 | 2:14700237-14700411 + | CCCCCACA-------CAC------ACAAAACTCGTGGCTATGAACTC-CTTCTCAGTGG-TAAGTAGGA------------AGACCAGA-GG--------A-----------------------CCGAC---------------------CAAGCGGATTGTGGAA----------------------------------------------CAATTGGATT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A-CCATTA-CCATCATTA-CCTTG-CATTGCAGTTCTTAGTGTTCGCCCTA--GCTGCGCT-------------GGCCGCTGCGGAGCCCC---CATCGGG | |
| droPer2 | scaffold_0:5714454-5714651 - | CCCCCACA-------CAC------ACAAAACTCGTGGCTATAAACTCCCTTCTCAGTGGATACGTAGGAT------------GACCAGA-GA--------ACCGACCTAGCCGGATT-------GTG-----------------------------------GGAA----------------------------------------------C------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTTGGATTA-CCATTAACAATCGTTATCCTTGCATTTGCAGTTCCTAGTGTTTCCCCTTATTCTGCGCTAGGCCTGTGTTGAGGCCCTAGCGGGGCTACAACTATCC-- | |
| droWil2 | scf2_1100000004943:8185262-8185319 + | A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTGCAGTTCTTAGTGTTCGCTCTG--GCCGCATT-------------GGCAGCTGCTGAGCCTC---CATCGGG | |
| droVir3 | scaffold_13047:11125080-11125137 - | CCCCCACA-------CATACAC-CACAAAACTCATTGCTATGAACTC-CTTCTCAGTGG-TAAGTAGC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droMoj3 | scaffold_6540:30634627-30634687 + | CTCG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCTACAGTTCTTAGTGTTTGCGCTC--GCTGCTCT-------------GGCCGCAGCTGAGCCGC---CATCGGG | |
| droGri2 | scaffold_14624:1011616-1011845 - | CCCAAACACCACACACACACACACACACAAA-TACGGCAATGAACTC-CTTCTCAGTGG-TAAGTTAAAC-TACAAC--AAGGATTACAGTG-GATAAA---------------------------------------------------TCAGTGGATTACAACCCCCTCA-----------------------------------------------ACC--------------C------------------------------------------------CCTCCTCTCGCTCT---C------------------------------------------------------------------------------TCTCTCTCCACTTTCCCACACTC------------------------------------------TCTCTACCCCCGT----TC---TCCACACAGTTCTTGGTATTTGCATTG--GCTGCTTT-------------GGCCGCAGCTGAGCCAC---CATCTGG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/19/2015 at 05:32 AM