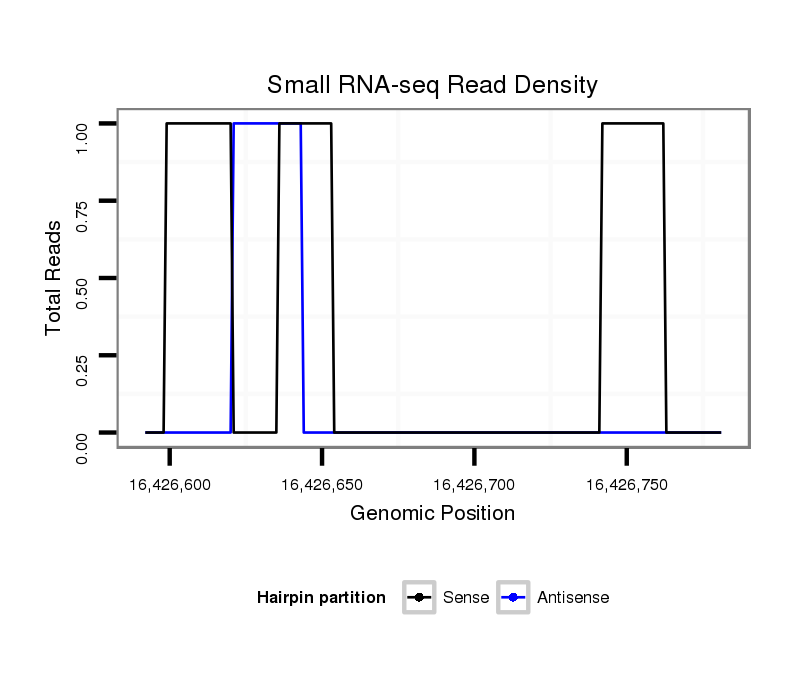
ID:dsi_13851 |
Coordinate:3l:16426642-16426731 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [3l_16426557_16426641_+]; CDS [3l_16426557_16426641_+]; CDS [3l_16426732_16426797_+]; exon [3l_16426732_16426797_+]; intron [3l_16426642_16426731_+]
No Repeatable elements found
| ##################################################------------------------------------------------------------------------------------------################################################## GGACGTGGTCACATCGTACCGCAGCTTGAGCTACGACCTGCAGAGTTTCGGTAAGCAACAGCTGATTAAAGCATCCCACCTGGCAAAGGTATTATTTATGTAAGTGCATCTATTATTTATACATTATTCTCTGCTTTTAGTGTTCAACGATGGCACTTCCAAGAAATTGTTCACCATACAGGGCACTATT **************************************************...........(((...((((((.............(((((.((...((((....)))).)).))))).............)))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
M053 female body |
SRR553485 Chicharo_3 day-old ovaries |
SRR902008 ovaries |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................AGCAACCGCTGATGAAAGCAG.................................................................................................................... | 21 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................TGGCACTTCCAAGAAATTGTT................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................GTTTCGGTAAGCAACAGC................................................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......GTCACATCGTACCGCAGCTTGA................................................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................TAGCAACCGCTGATGAAAGCA..................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................TCACCTGCAGAGTCTCGGTA......................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................TTAAGCATCCCGCCTGG........................................................................................................... | 17 | 2 | 10 | 0.20 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .............................GCTACTACCAGCAGAGT................................................................................................................................................ | 17 | 2 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................GCTACGATCTGCTGAGT................................................................................................................................................ | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........CATCTTACCGCAGCATTAG................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
CCTGCACCAGTGTAGCATGGCGTCGAACTCGATGCTGGACGTCTCAAAGCCATTCGTTGTCGACTAATTTCGTAGGGTGGACCGTTTCCATAATAAATACATTCACGTAGATAATAAATATGTAATAAGAGACGAAAATCACAAGTTGCTACCGTGAAGGTTCTTTAACAAGTGGTATGTCCCGTGATAA
**************************************************...........(((...((((((.............(((((.((...((((....)))).)).))))).............)))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR902009 testis |
M023 head |
M025 embryo |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................ATCGTTGGCGACTACTTTCGT..................................................................................................................... | 21 | 3 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................TGGACGTCTCAGAGCCAT......................................................................................................................................... | 18 | 1 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................AGACGAAAGTCACAAGTTGCT........................................ | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................TCGATGCAGGACGTCTTAAAG............................................................................................................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................TCGTTGGCGACTACTTTCGTC.................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................CGATGCTGGACGTCTCAAAGCCA.......................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................ACAGGGAAGGTTCTTTAA...................... | 18 | 2 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ....................................GGACGTCTCAGAGCCAT......................................................................................................................................... | 17 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................ACGTCTCAGAGCCAT......................................................................................................................................... | 15 | 1 | 9 | 0.22 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................AGTGGACGTCTCAGAGCCAT......................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................TCGAACTCGATGCAG......................................................................................................................................................... | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................AGTTGCTCCCGTTCAGGTTCT.......................... | 21 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................AGACGTCTCAGAGCCAT......................................................................................................................................... | 17 | 2 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................AGTGGACGTCTCAGAGCCA.......................................................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................TCCGACTCGATGCTGGGC...................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................AACTTGATGCTGGAGGTCC.................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 04:51 AM