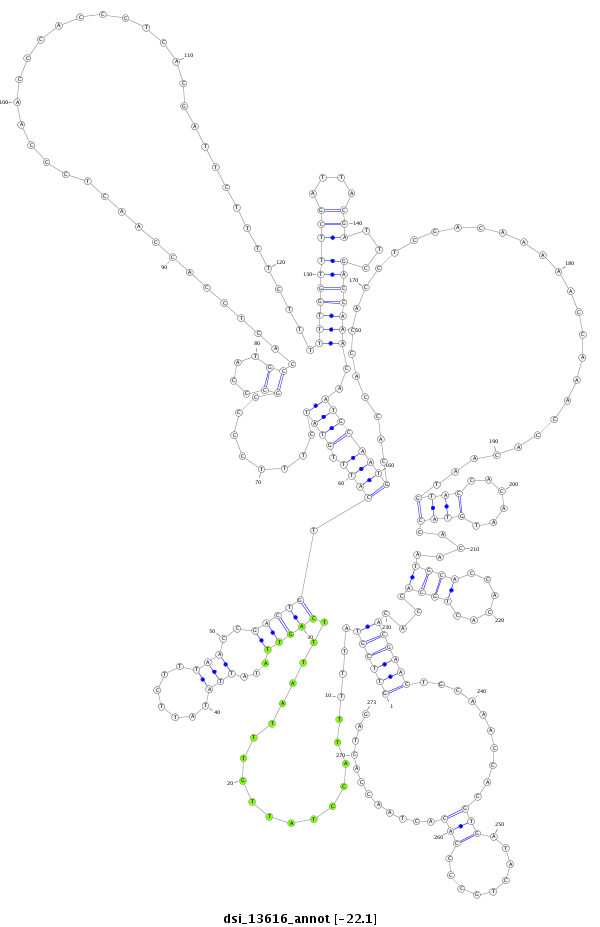
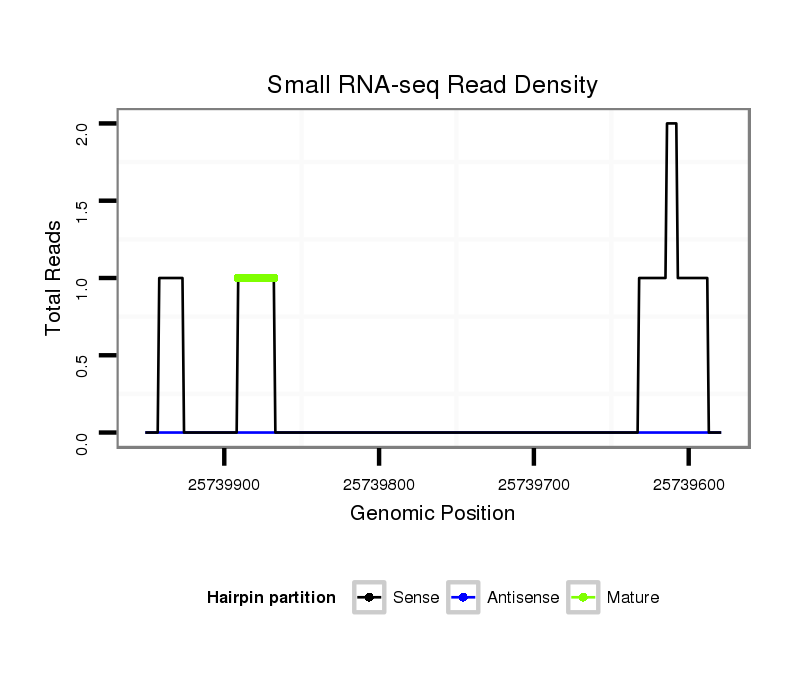
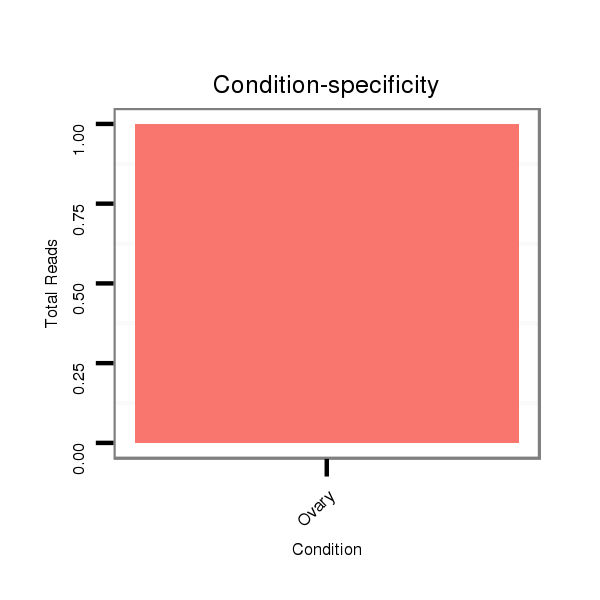
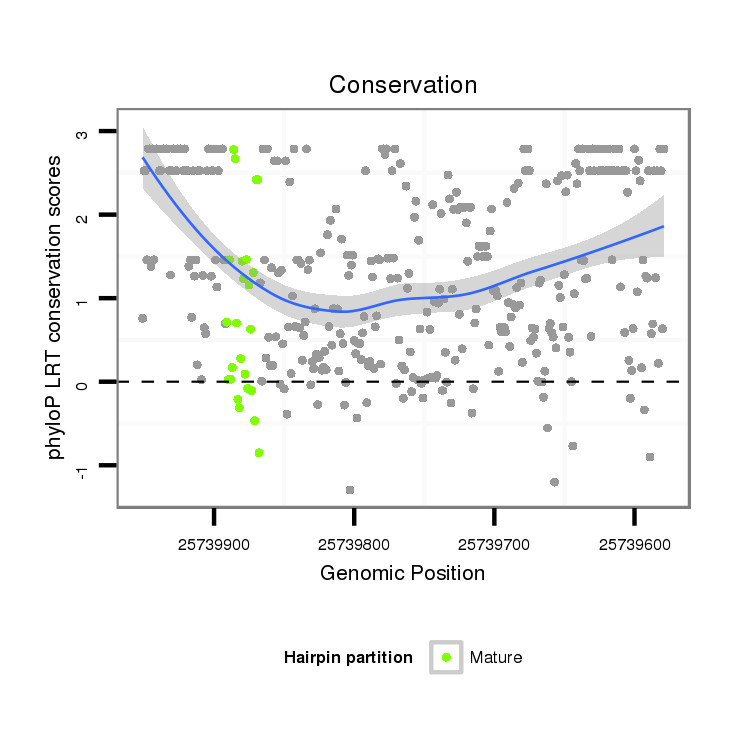
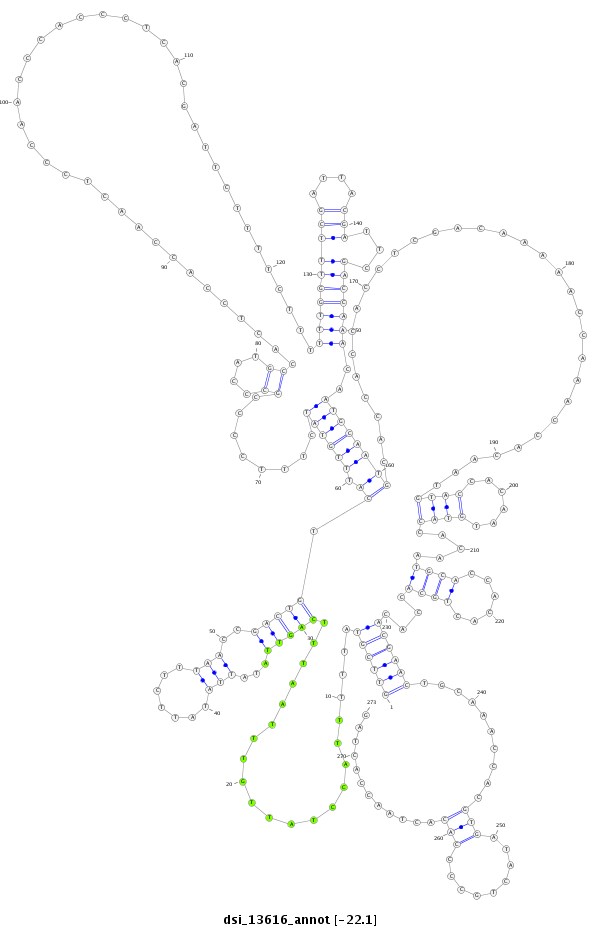
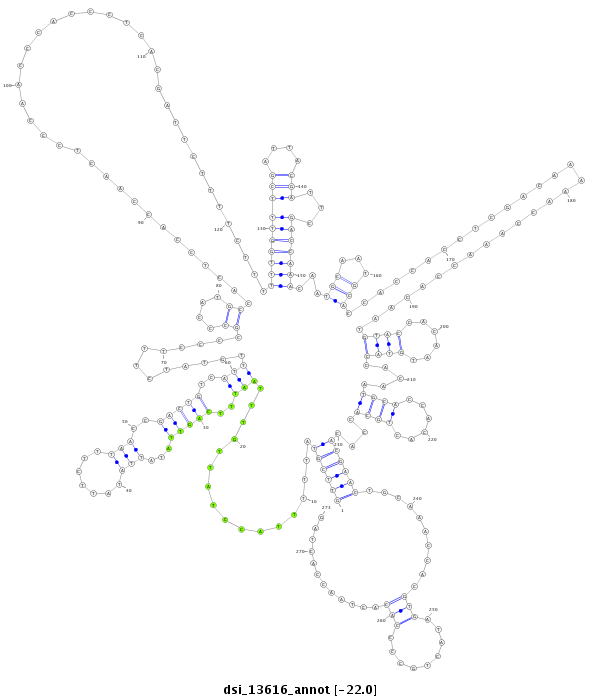
ID:dsi_13616 |
Coordinate:3r:25739629-25739901 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -22.1 | -22.1 | -22.0 |
 |
 |
 |
exon [3r_25739902_25739958_-]; CDS [3r_25739902_25739958_-]; exon [3r_25739484_25739628_-]; CDS [3r_25739484_25739628_-]; intron [3r_25739629_25739901_-]
| Name | Class | Family | Strand |
| (GTGTG)n | Simple_repeat | Simple_repeat | + |
| ##################################################---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AGCAATTTATTAGGAAATTCAGTTGAAATGTGCTTACCAGCGACGCCTAGGTTCGTATTTTTACCTATTGTTTAATTTCAGTTATATTATATTCTTTAACCGACTGTCATTTGTATCTTTCCCCGCCCATGCCACTCCACCAACTCCCAACCCACCCTCACGATTCTTTTCTTTTTTGGTTTCGATTACGATTCGACCAAACAATGCAATGCACCACCACCTCGACAAAAACCAAACCACAATGTACCACAATGTACCACAATGCACCACACTGCACCACACGAACTGCAAACCACGTGATACTGCCCCACACTAACCACTAGAAACTCTACTCCACAACACGCCCCAACCAGCAGCGCCCGCCAGCGAACCA **************************************************((((((......................(((((...(((.......)))..))))).((((.((((........((....))..........................................((((((((((....)))...)))))))..))))))))................................((((......)))).....((((......))))....))))))..........(((.........)))............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M025 embryo |
SRR902008 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................................................................................................................................AACACGCCCCAACCAGCAGCGCCCGCC......... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........TTAGGAAATTCAGTTG............................................................................................................................................................................................................................................................................................................................................................ | 16 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................TTACCTATTGTTTAATTTCAGTTA................................................................................................................................................................................................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................CTAGAAACTCTACTCCACAACACGC............................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................TAACGGACTGTCATTTTTAACTT.............................................................................................................................................................................................................................................................. | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........TGAGGAAATTCAGTTGAAATATGA.................................................................................................................................................................................................................................................................................................................................................... | 24 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................CCCACCCTCACCATT................................................................................................................................................................................................................ | 15 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........TGAGGAAATTCAGTTGAAATT....................................................................................................................................................................................................................................................................................................................................................... | 21 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................TCTTTCACCGCCCGTGTCA............................................................................................................................................................................................................................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................AACCCCCCCTCACTATT................................................................................................................................................................................................................ | 17 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................ATTCCAGTTGTATTATAATC....................................................................................................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................................................................TAGCACCACCTTGACAAAA............................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
TCGTTAAATAATCCTTTAAGTCAACTTTACACGAATGGTCGCTGCGGATCCAAGCATAAAAATGGATAACAAATTAAAGTCAATATAATATAAGAAATTGGCTGACAGTAAACATAGAAAGGGGCGGGTACGGTGAGGTGGTTGAGGGTTGGGTGGGAGTGCTAAGAAAAGAAAAAACCAAAGCTAATGCTAAGCTGGTTTGTTACGTTACGTGGTGGTGGAGCTGTTTTTGGTTTGGTGTTACATGGTGTTACATGGTGTTACGTGGTGTGACGTGGTGTGCTTGACGTTTGGTGCACTATGACGGGGTGTGATTGGTGATCTTTGAGATGAGGTGTTGTGCGGGGTTGGTCGTCGCGGGCGGTCGCTTGGT
**************************************************((((((......................(((((...(((.......)))..))))).((((.((((........((....))..........................................((((((((((....)))...)))))))..))))))))................................((((......)))).....((((......))))....))))))..........(((.........)))............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR553487 NRT_0-2 hours eggs |
M024 male body |
SRR618934 dsim w501 ovaries |
M025 embryo |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR902009 testis |
SRR553488 RT_0-2 hours eggs |
M053 female body |
SRR902008 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................................CTGTTACCTGATGTTACGTG.......................................................................................................... | 20 | 3 | 13 | 18.00 | 234 | 147 | 9 | 27 | 7 | 23 | 7 | 5 | 1 | 3 | 5 | 0 |
| ........................................................................................................................................................................................................................................................TGTTACCTGATGTTACGTG.......................................................................................................... | 19 | 2 | 4 | 3.75 | 15 | 7 | 3 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................................................................................................................................................................GTTACCTGATGTTACGTG.......................................................................................................... | 18 | 2 | 8 | 3.63 | 29 | 20 | 0 | 8 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................TTGGGTGGGAGTGCTGAGAAA............................................................................................................................................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................AAAGGCGCGGGTACGGTGACGT.......................................................................................................................................................................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................AAAAGGCGCGGGTACGGTGACGT.......................................................................................................................................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................ACGGTGAGATGGTTGA.................................................................................................................................................................................................................................... | 16 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................TGGTCCCTGCGGATCC.................................................................................................................................................................................................................................................................................................................................. | 16 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................TGGCCCCTGCGGATCC.................................................................................................................................................................................................................................................................................................................................. | 16 | 2 | 8 | 0.75 | 6 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ...................................TGGCCCCTGCGGATCCA................................................................................................................................................................................................................................................................................................................................. | 17 | 2 | 3 | 0.67 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................CGGTCTTGTGCGGGGTTG....................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................CGGTGAGATGGTTGA.................................................................................................................................................................................................................................... | 15 | 1 | 5 | 0.40 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................TACTGCTCAGCTGGTTTGT.......................................................................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................AAGGCGCGGGTACGGTGACGT.......................................................................................................................................................................................................................................... | 21 | 2 | 6 | 0.33 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................GATTGGTGATCTGTGCGA........................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................AAGAAATTGTCTGACA.......................................................................................................................................................................................................................................................................... | 16 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................CAGGTGTTGTTCGGGGTCGGT..................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................TGTGACGTGGTGAGCTAGAGGT................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................TGAGACGAGGTGTTGT................................ | 16 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................ACGGTGAGATGGTTGAC................................................................................................................................................................................................................................... | 17 | 2 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................GTTACGTGATGTTACGTG.......................................................................................................... | 18 | 2 | 14 | 0.14 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................GTTACGTGATGTTACGTG................................................................................................ | 18 | 2 | 14 | 0.14 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................CTGTGAGATGAGGTGT................................... | 16 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................GGTGAGGGTTGGGTGG......................................................................................................................................................................................................................... | 16 | 1 | 15 | 0.13 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................CTGTTACTTGATGTTACGTG.......................................................................................................... | 20 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................GGGGTAGGATTGGTGATCTG................................................ | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................AAGGCTCGGGTACGGTGACGT.......................................................................................................................................................................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................TTTTGCGGGGTTGGTCTT.................. | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................GGTGATCTTTGAGAGTA........................................ | 17 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................CCAAAGCCCATGCTAAG................................................................................................................................................................................... | 17 | 2 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................TGGATAACAAATTGA........................................................................................................................................................................................................................................................................................................ | 15 | 1 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................TGATTGGTGGTCTGTGACA........................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................GGCCCCTGCGGATCCA................................................................................................................................................................................................................................................................................................................................. | 16 | 2 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................CTGTGAGATGAGGTG.................................... | 15 | 1 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................TGGTATGCTTGACGT................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........ATCCTTTATGTCAACTA.......................................................................................................................................................................................................................................................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................ACGTAGAAAGGGGCGATTA................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................GGCCCCTGCGGATCC.................................................................................................................................................................................................................................................................................................................................. | 15 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................TGGTCGCTGCGGTTACAAC............................................................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................ACGGTACGTGGTGGTGG........................................................................................................................................................ | 17 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3r:25739579-25739951 - | dsi_13616 | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTTACCAGCGACGCCTAGGTTCGTATTTTT---ACC------------TATTGTTTA-------------------------------ATTTCAGTTATA-TT---------------A-------TAT-TCTTTAACCG------ACTGTCATTTG--------TATCTTT-CCC---CG---------------------------------------------------------CCCATGCCACTCCACC-AACT-CCCAACCC----A-----------------CC--C--TCACGATTCT-TTTCTTTTTTGGTTTC------GATTACG------ATT----CGACCAAAC-AATGCAATGCACC---------ACCACCT-----CGACAA-------AAACCAA---ACCACAA-TG-TACCACAA-------TGTACCACA--------------------A-------------------------------------------------------------------TGCACCACACTGCA--CCACACGAACTGC---------------AAAC--------------------------------------------------CACGTGATAC--TGCC-CCACACTAACCACT-----------AGAAACTCTACTCCACAACACGCCCCAAC---CAGCAGCGCCCGCCAGCGAACCA |
| droSec2 | scaffold_4:5246452-5246809 - | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTTACCAGCGACGCCTAGGTTCGTATTTTT---ACC------------TATTGTTTA-------------------------------ATTTCAGTTTAA-TT---------------A-------TAT-TCTTTAACCG------ACTGTCATTTG--------TATCTTT-CCC---CG---------------------------------------------------------CCCATGCCACTCCACC-AACT-CCCAACCC----A-----------------CC--C--TCACGATTCT-TTTCTTTTTTGGTTTC------GATTACG------ATT----CGACCAAAC-AA-----TGCACC---------ACCACCT-----CGACAA-------AAACCAA---ACCACAA-TG-TACCAC-----------------A--------------------A-------------------------------------------------------------------TGCACCACACTGCA--CCACACGAACTGC---------------AAAC--------------------------------------------------CACGTGATAC--TGCC-CCACACTAACCACT-----------AGAAACTCTACTCCACAACACGCCCCAAC---CAGCAGCGCCCGCCAGCGAACCA | |
| dm3 | chr3R:26419522-26419884 - | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTTACCAGCGACGCCTAGGTTCGTATTTTA---AAC------------TATTGTTTA-------------------------------ATTTCAGTTTAA-TT---------------G-------TAT-TCTTTAACCG------ACTGTCATTTG--------TATCTTT-CCC---CA---------------------------------------------------------CCCATGCCACTCCACC-AACT-CCCAACCC----A-----------------CC--C--TCACGATTCT-TTTCTTTTTTGGTTTC------GATTACG------ATT----CGACCAAAC-AATGCAATGCACC---------ACCACCT-----CGACAA-------AAACCAA---CTCACAA-TG-TACCAC-----------------A--------------------A-------------------------------------------------------------------TGCACCACACTGCA--CCACACGAACTGC---------------AAAC--------------------------------------------------CACGTGATAC--TGCC-CCACACTAACCACT-----------AGAAACTCTACTCCACAACACGCCCCAAC---CAGCAGCGCCCGCCAGCGAACCA | |
| droEre2 | scaffold_4820:1582141-1582517 + | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTTACCAGCGACGCCTAGGTTCGTATTTTA---AAC------------TATATTTTA-------------------------------ATTTCAGTTTAA-TT---------------G-------TAT-ACTTTATCCG------ACTGACATTTG--------TATTTTT-CCC--CCA-----------------------------------CCCATGCCACCC----ATGCCACCCATGCCACTCCACC-AACT-C-----CC----A-----------------CC--C--TCATGATTCT-TTTCTTTTTTGGTTTC------GAATACG------ATT----CGACCAAAC-AATGCAATGCACC---------ACCACCT-----CGACAA-------AAACCAA---CCCACAA-TG-TACCAC-----------------A--------------------A-------------------------------------------------------------------TGCACCACATTGCA--CCACACGAACTGC---------------AAAC--------------------------------------------------CACGTGATAC--TGCC-CCACACTAACCACT-----------AGAAACTCTACTCCACAACACGCCCCAAC---CAGCAGCGCCCGCCAGCGAACCA | |
| droYak3 | 3R:27339744-27340125 - | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTTACCAGCGACGCCTAGGTTCGTATTTTT---AAC------------TATTGTTTA-------------------------------ATTTCAGTTTAA-TT---------------A-------TAT-TCTTTAACCG------ACTGTCATTTG--------TATTTTT-CCC--CCA------------------------------------AA-------------------ACCATGCCACTCCACC-AACT-CCCAACCC----A-----------------CC--C--TCATGATTCT-TTTCTTTTTTGGTTTC------GAATACGAATGCGATT----CGACCAAAC-AATGCAATGCACC---------ACCACCT-----CGACAA-------AAACCAA---CCCACAA-TG-TACCACAA-------TGTACCACA--------------------A-------------------------------------------------------------------TGCACCACATTGCA--CCACACGAACTGC---------------AAAC--------------------------------------------------CACGTGATAC--TGCC-CCACACTAACCACT-----------AGAAACTCTACTCCACAACACGCCCCAAC---CAGCAGCGCCCGCCAGCGAACCA | |
| droEug1 | scf7180000409555:926076-926449 - | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTTACCAGCGACGCCTAGGTTCGTATTTTT---AAC------------TATTGTTTC-------------------------------ATTTCAGTTCAA-TT---------------A-------TAA-TTTTTAATCA------ACTGCCATTTG--------TACTTTT-CCC---CA-----------------------------------CCCA-------------------CCACGACACACCACC-AACC-CCCGACCC----A-----------------CC--C--ACACGATTTC-TTTCTTTTTTGGTTAC------GATTACG------ATC----CGACCATCC-AATGAAACGCACCACAACCACCACCACCT-----CGACAA--------AACCAA---TCCACAA-TG-TACCAC-----------------A--------------------A-------------------------------------------------------------------TGCACCACATTGCA--CCACTCGAACTGC---------------AAAC--------------------------------------------------CTCGTGAAAC--TGCC-GCACACTAACCACT-----------AGAAACTCTACTCCACAACACGCCCCATC---CAGCAGCAACCGCCAGAGAACCA | |
| droBia1 | scf7180000302113:941127-941515 + | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTTACCAGCGACGCCTAGGTTCGTATTTTT---AAC------------TATGGTTTC-------------------------------CATTCAGTTAAG-TT---------------A-------TAA-TTTTTAATCA------ACTGCCATTTG--------TACTTTT-CCC---CAC--------------------CTCCCACACCCACAC---------CC----ACG------CCGCCACTCCACC-CACTTCCCCACCC----G-----------------CC--C--ACATGATTCT-TTTCTTTTTTGGTTTC------GATTACG------ATC----CGACCAACC-AATGAAACGCACC---------GCCACCT-----CGACAAAACGACAAAACCAA---CCCACAA-TG-TACCAC-----------------A--------------------A-------------------------------------------------------------------TGCACCACATTGCA--CCACTCGAACCGC---------------AAAC--------------------------------------------------CACGTGAAAC--TGCC-CCACACTAACCACT-----------AGAAACTCTACTCCACAACACGCCCCATC---CAGCAGCACCCGCCAGCGGACCA | |
| droTak1 | scf7180000415367:378353-378731 + | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTTACCAGCGACGCCTAGGTTCGTATTTTT---AAC------------TATTGTTTC-------------------------------ATTTCAGTTTAA-TT---------------A-------TAA-TTTTTAATCA------ACTGCCATTTG--------TACTTTTTC-C--CCA-----------------------------------CCCAC-CCAACC----ACG------CCGCCACTCCACC-AACT-TCCAACTTC---CCCAAA-ATCCACCACCACCCAC--ACATGATTCT-TTTCTTTTTACGATCC------GATC----------------TGAACTA-A-T-----CTGCACA---AC---CACCACAT-----CGACAA--------AACCAA---ACCACAA-TG-TACCAC-----------------A--------------------A---------A---------------------------------------------------------TGCACCACATTGCA--CCAATC------C---------------GAAC--------------------------------------------------CGCGTGAAAC--TGCC-GCACACTAACCACT-----------AGAAACTCTACTCCACAACACGCCCCAAC---CAGCAGTAACCGCCAGCGGACCA | |
| droEle1 | scf7180000491280:2747648-2748057 - | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTTACCAGCGGCACCTAGGTTCGTATTTTT---AAC------------TATTGTTTA-------------------------------ATTTTAGTTAAA-TT---------------A-------TAATTTTTTAATCA------ACTGCCATTTG--------TACTTTTT-TTCCCCA-----------------------------------ACAA------CC----ACG-------CGCCACTCCACCAAATTTCCCCACCC----A-----------------CC--C--ACATGATTCT-TTTCTTTTTTGGTTTC------GATAACG------ATT----CGACC-----AATGAAATGCACC---------ACCACTT-----CTACAA-------AAACCAA---CCCACAA-TG-TACCACAA-------TGTACCACAATGTACCACAATGTACCACAATGCACCACAT---------------------------------------------------------TGCACCACATTGCA--CCACTCGAATCAC---------------AATC--------------------------------------------------CTCGTGAAAC--TGCC-GCACACTAACCACT-----------AGAAACTCTACTCCACAACACGCCCCAAC---CAGCAGCACCCGGCAGCGAACCA | |
| droRho1 | scf7180000778339:12730-13099 + | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTTACCAGCGGCGCCTAGGTTCGTATTTTT---AAC------------TATTGTTTC-------------------------------ATTTCATTTAAT-TT---------------A-------TAA-TTTTTAATCA------ATTGACATTTG--------TACTTTT-TTC---CA-----------------------------------CCAA------------------CCCACGCCACTCCACA-AATACCCCAACCC----A-----------------CC--C--ACATGATTCT-TTTCTTTTTTGGTTTC------GATAACG------GTT----CGACCAACC-AATGAAATACACC---------ACCA-------------A-------AAACCAA---CCCACAA-TG-TACCACAA-------TGTACCACA--------------------A-------------------------------------------------------------------TGCACCACATTGCA--CCACTCGAACCAC---------------AAAC--------------------------------------------------CTCGTGAAAC--TTCC-GCACACTAACCACT-----------AGAAACTCTACTCCACAACACGCCCCAAC---CAGCAGCAACCGGCAGCGAACCA | |
| droFic1 | scf7180000454106:52035-52398 + | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTTACCAGCGGCGCCTAGGTTCGTATTTTT---AAC------------TATTGTTTC-------------------------------ATTTCAGTTTAA-TT---------------A-------TA--TTTTTAATCA------ACCGCCATTTG--------TACTTTT-CCC--CCA-----------------------------------CCA-------------------ACCACGCCACTCCACC-AACT-CCCCACCC----A-----------------CC--C--ACATGATTCT-CTTCTTTTCTGGTTTC------GATTACG------ATC----CGACCATCC-AACG-AATGCACC---------ACCACCT-----CGACCA-------AAACCAA---CCCACAA-TG-TACCAC-----------------A--------------------A-------------------------------------------------------------------CGTACCACATTGCA--CCTCTCGAACTGC---------------G-AC--------------------------------------------------CACGTGAAAC--TGCC-GCACACTAACCACT-----------AGAAACTCTACTCCACAACACGCCCCATC---CAGCAGCAACCGCCAGCGGACCA | |
| droKik1 | scf7180000302390:388115-388458 - | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTTACCAGCGATGCCTAGGTTCGTATTTTTTTAAAC------------TATTGTTTCA------------------------------ATTTCAGTTAAATTT---------------A-------TGA-TTTTTAATCGAA----AATGCCATTTG--------TACTTTTTC-C--CC-CCGC----------CAC----CT--------------------------------------------CCCACA-AAC---CCCACCC----A-----------------CC--C--ACACGATTCT-TCTCTTTTTTGGTTTAC-----GATTACA------ATC----CGAA---ATATACGAAATGCACC---------AACACCT-------------------------------------------------------------------------------------------------------------------------------------------------------TGCACCACATTGCACACCACTCGAACCAC---------------GATC--------------------------------------------------CATGTGAAACTCTGCTGCCACACTAACCACT-----------AGAAACTCTACTCCACAACACGCCCCGATCAGCAGCAGCAACAGGCAGCGCACCA | |
| droAna3 | scaffold_12911:4562867-4563229 + | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTTACCAGCGACGCCTAGGTTCGTATTTTT---ACC------------TATTGTTTC-------------------------------ATTTCAGTTAAA-TT---------------A-------TGA-TTTTTAATCA------ATTGCCATTTG--------TACTTTTTC-C--CCACCGCTCACCAAATCCACCCAC------------------------CCCCAAAAACCACTCACTCCACTCCAC--------------C----A-----------------CC--C--ACATGATTCT-TTTCTTTTTTGGTTTC------GATTACG------ATC-CAATGACCA-----------------------------ACCT-----CGACAA--------AACCAACAACACT------------C-----------------A--------------------A-------------------------------------------------------------------TGCACCACATTGCA--CCACTCGAAATAC-------------CTAAAC--------------------------------------------------CGCGTGAAAC--TGAC-GCACACTAACCACT-----------AGAAACTCTACTCCACAACACGCCCCATC---CAGCAGCAGCCGGACGCGCACCA | |
| droBip1 | scf7180000396640:1405706-1406077 + | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTTACCAGCGACGCCTAGGTTCGTATTT-T---ACC------------TATTGTTTC-------------------------------ATTTCAGTTAAA-TT---------------A-------TGA-T-TTTAATCA------ATTGCCATTTG--------TACTTTTTC-C--CCACCGT------AATCCAC-CAC------------------------CCCAAAATACCACTCACTCCACTCCACC-------------C----ACCCAACAACCACCCACACC-AC--ACATGATTCT-TTTCTT-TTTGGTTTC------GATCTCG------ATC----C---------AA-----TGAA------------CAACCT-----CGACAA--------AACCAACAACACT------------C-----------------A--------------------A-------------------------------------------------------------------TGCACCACATTGCA--CCACTTGACACAC-------------TTGAAC--------------------------------------------------CGCGTGAATC--TGAC-GCACACTAACCACT-----------AGAAACTCTACTCCACAACACGCCCCATC---CAGCAGCAACCGGACGCGCACCA | |
| dp5 | 2:820094-820401 + | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTTTCCAGCGGCGCCTAGGTTCGTATTATTTT-ATT------------TATT---TG------------------------ATTATTCATTCGAATTAAT-TT---------------A-------TGA-TTTCTAATCG------ATNNCCACTCAACCCCCCACCCTTTCTC-C--CCA------------------------------------------------------------------CTCC-CC-AACC------CCC----G-----------------CC--C--ACACGGTTCTGTTTCTTTTTTGGTTACCAACAC----ACG------A----------------------------------------------------------------------CACCACA------------C-----------------A-------------------------------------------------------------------------------------------------CTGCA--CCACA----------ATCGAC---CCTCGGACC-------------------------------------------------CTCG-----------------ACCAACCACTCTAATCCGTAAAGAAACTCTACTCCACAACACGCACCCATTAGCAGCAGCAGCCGACAGCGCACCA | |
| droPer2 | scaffold_2942:101-409 + | AGCAATTTATTAGGAAATT---CAGTTGAAATGTGCTGTCGAGCGGCGTCTACGTGCGTATTTTTTT-ATT---------------TATTTG------------------------ATTATTCACTCGAATTAAT-TT---------------A-------TGA-TCTCTACTCG------ATTGCCACTCAACCCCCCATCCTTTCTC-C--CCA------------------------------------------------------------------CTCC-CC-AACC-TC----CG----T-----------------CC--G--ACACTGTTCTGTTTCTTTTTTGGTTACCTACAC----ACG------A----------------------------------------------------------------------CACCACA------------C-----------------A-------------------------------------------------------------------------------------------------CTGCA--CCACA----------ATCGAC---CCTCGGACC-------------------------------------------------CTCG-----------------ACCAACCACTCTAATCCGTAAAGAAACTCTACTCCACAACACGCACCCATCAGCAGCAGCAGCCGACAGCGCACCA | |
| droWil2 | scf2_1100000004943:9926584-9927008 - | AGCAATTTATTAGGAAATT---CTGTTGAAATGTGCTTACCAACGACACCTAGGTTTGTATTGAT---ATA------------TATACAT----------------ATATATATTA-------AGAGTAC----T-TCTCAATTGGTATTTATATTTTGTTT-A-TATTTAATTA------AATG-CATTTA--------TCTTTTT-TCA--TTT----------------------------------------------------------TCTT-TCCCTTTAT--------------CC---CCCAAA-AAAAAAAAAAAAAAATATATACGTT--T-ATCTTTTTTTGGTTAC------GAATGCG------A---------C------AACAAAATACTCA---AC---CACAACAACCACTCAAC----------------------------------------------------------------------------------------------------------------------------------------------TACACC--ACTGCA--CTCCACACATTGCC-ATCGATAACGTCTGCACCGTGCACTATGCAAAAAAATATTAAATAATAATAAATCAAAACAAATAAACCTGTCATACTTTG-T-CCACACTAACCACA-----------AGAAACTCTACTCCACAACATGCCCC------TAGCAGTAGCCGTACGCGCACTA | |
| droVir3 | scaffold_13047:11188301-11188685 - | dvi_16460 | TGCAATATATTAGGAAATT---CTGTTGAAATGTGCATACCATCGGCGTCTAGGTTTGTATTTTA---AATAC-------AAAT-ATGTAAC-------------------------------A---CTGTTAAT-TT---------------GTT-----TAG-TTTATACTGA------AACGCGAATTG--------TATATGT-AC-----------------------------------------------------------------------------------------------------------------------------TGTCATT-TTTCTTTTTTGGTTTC------AATATCG------ACAACTACGAC-----------AACGCACT---------ACCACAT--CCACAACAA-------CAAC-AACA-ACAACAATTGCTACTACAACAACAATTGC---------TA-------CTACAACAACGCACCACACCACAAAACATCTGCACACGCC--------------------ATCCAC---ACACAAC-----TACTCTGCA--CAAT--AAAATGCCA------------AAAAA--------------------------------------------------AATGTCAAA---TGTG-CCACACTATTAACC-----------AGAAACTCTACTCCACAACATGCAC---C---CAGTAGTAGCCGCCAGCGCACCA |
| droMoj3 | scaffold_6540:30566881-30567260 + | AGCGATATTTTAGGAAATT---CTGTTGAAATGTGCATGCCAGCGGCGTCTAGGTTTGTATTTTA---TATACAAAATATAAATATGTGTACATTTGCATATATGTATATG-CTCG-------ATATT-CTTGAC-TT---------------GTG-----TG------------------------AATTG--------TTTATGT-ATT---------------------------------------------------------------------------------------------GTCA-----------------TT--T--TTATTATTC--TTTATTTTTTGGTTAC------TACAACG------AAC----C-----------------ACACC---------AATACCT-----CAACAA-------CAAC-AACA----------------AC-----------------A--------------------ACGCACTACACTACA-TACATTTGCACATGTCAATTCGACACCACCAACACCACCCACACCACACAAC-----CACTCTGCA--C---ATTAAATGCTC--------------A-A--------------------------------------------------TATGCC-AAC--TACG-CGACACTAATCACA-----------AGAAACTCTACTCCGCAACATGCAC---C---CAGTAGCAGCCGTCAGCGCACCA | |
| droGri2 | scaffold_14624:1096849-1097168 - | TGCAATATATTAGGAAATTCTTCTGTTGAAATGTGCATACCGACGGCGTCTAGGTTTGTATCTACCTAAAC-------ATAAATACATATAC---TATATATATATATATAATTTG-------ACATTAT----T-TT---------------GTG-----TGT-TTTATTTTCGAATATAATTGCGAACTG--------TATATGT-GCT---------------------------------------------------------------------------------------------ATC-----------------------------ATTTTC-TTTATTTTTTGGTTAC------GATT----------------TGAACAA-C-TCTGCAA---------------------------------------------------------------------------------------------------------CTGCACCACA----------------------------------------------------------CACATCAATATGCA--CCACATCAAATGC---------------CAAT--------------------------------------------------TG---CAAAC---GCTTACACACTAATCACA---------ATAGAAACTCTACTCCACAACATGCACCCAT---TAGC---AGTCGGCAGCGCACTA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 01:04 PM