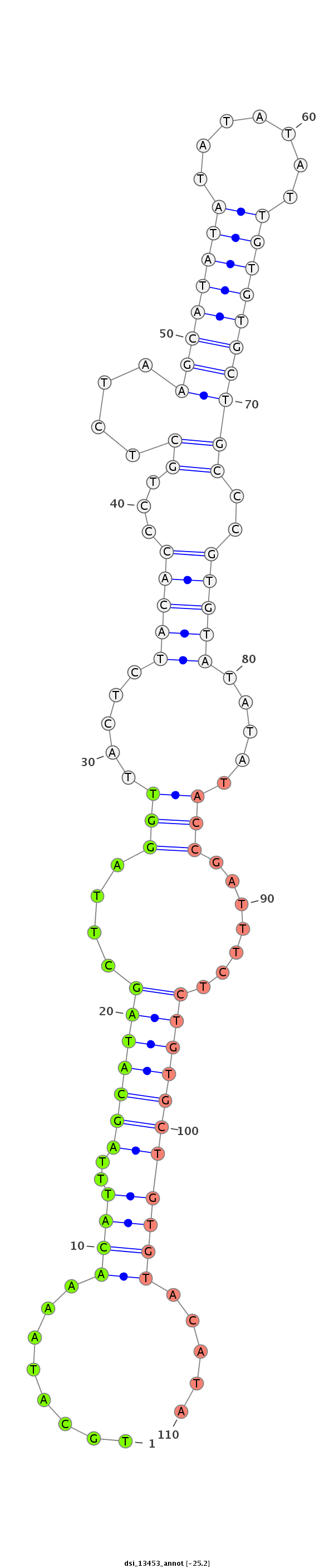
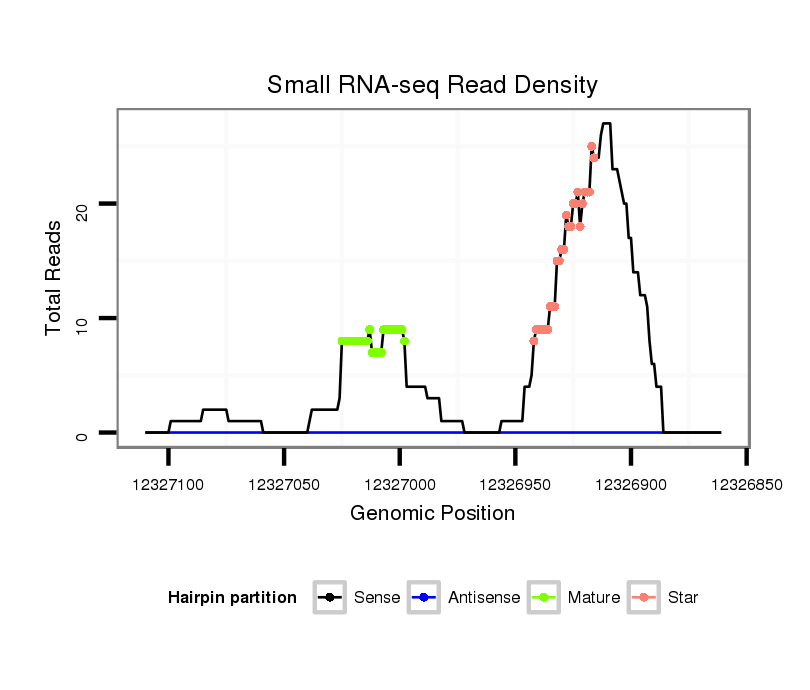
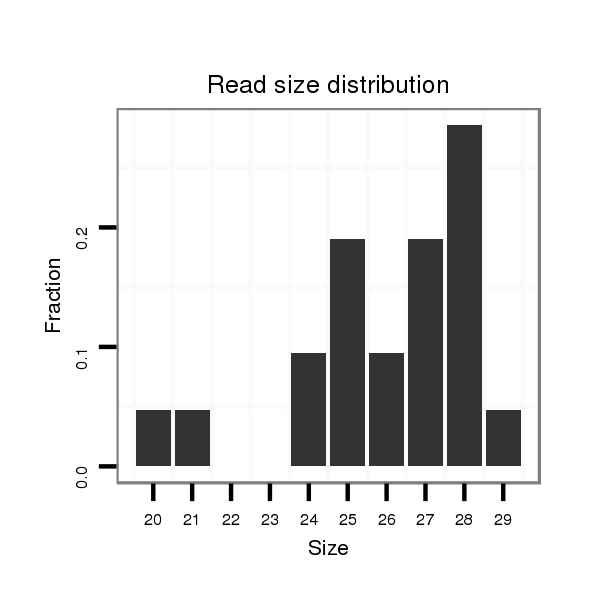
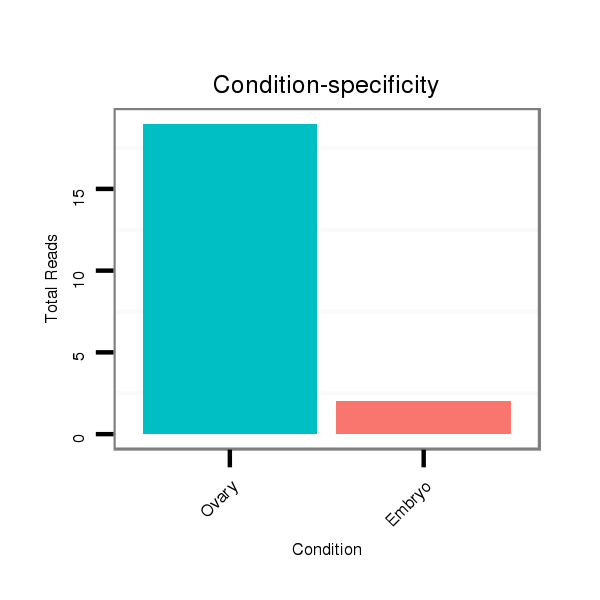
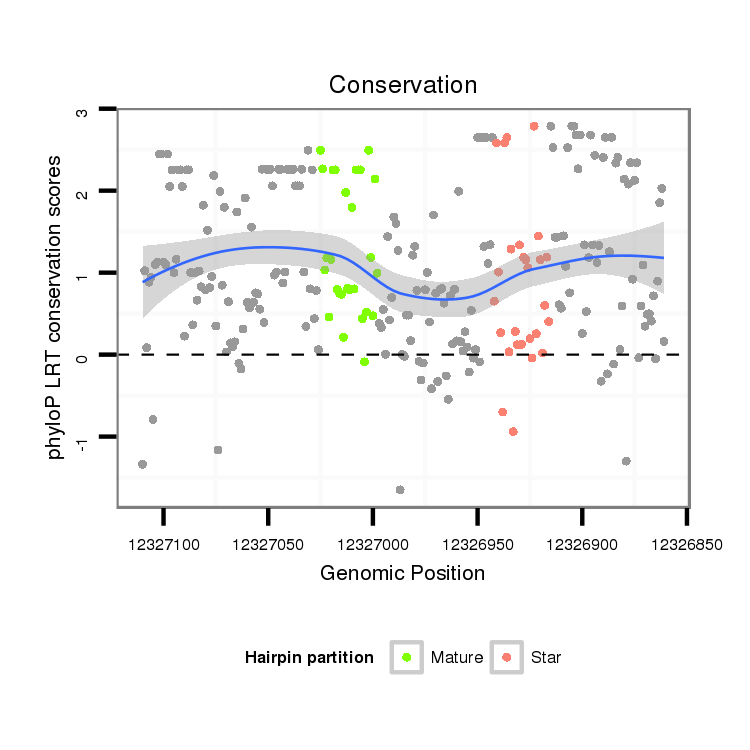
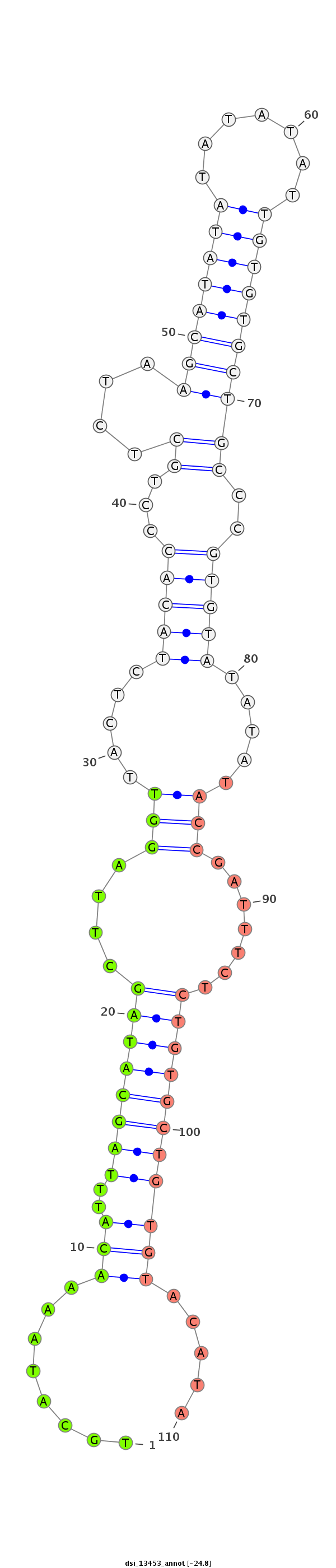
ID:dsi_13453 |
Coordinate:2r:12326911-12327060 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -24.8 | -24.2 |
 |
 |
intergenic
No Repeatable elements found
|
GTGGCAAAATTTACTAAAACCATTTTCTGCTCGTTATACCCCGCCCACGTGAGCAACTAAATGAAAACTATTTGGCAAATGTGGCTGCATAAAACATTTAGCATAGCTTAGGTTACTCTACACCCTGCTCTAAGCATATATATATATTGTGTGCTGCCCGTGTATATATACCGATTTCTCTGTGCTGTGTACATATGTAGGTCGTAATGTAAGGTACCAACAACAACAGCAGCAGCAATACAACAATCAA
*************************************************************************************........((((..(((((((....(((.....(((((...((....((((((((.......))))))))))..))))).....))).......))))))))))).....******************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
M025 embryo |
SRR553487 NRT_0-2 hours eggs |
M024 male body |
SRR618934 dsim w501 ovaries |
O002 Head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................TGCATAAAACATTTAGCATAGCTTAGGT......................................................................................................................................... | 28 | 0 | 1 | 5.00 | 5 | 1 | 4 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................TGCTGTGTACATATGTAGGTCGTAATG......................................... | 27 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................TGTGTACATATGTAGGTCGTAATGTA....................................... | 26 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................TCTGTGCTGTGTACATATGTAGGT................................................ | 24 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................TAGGTCGTAATGTAAGGTACCAACAAC.......................... | 27 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................TACATATGTAGGTCGTAATGTAAGG.................................... | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TATGTAGGTCGTAATGTAAGGTACC................................ | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................TACCGATTTCTCTGTGCTGTGTACATA....................................................... | 27 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................TAGCTTAGGTTACTCTACACCCTGC.......................................................................................................................... | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................TATATACCGATTTCTCTGTGCTGT.............................................................. | 24 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................TTCTCTGTGCTGTGTACATATGTAGGT................................................ | 27 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................ACCTCAGCAGCAATACAACAATCAA | 25 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................TATATACCGATTTCTCTGTGCTGTGTA........................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................CATATGTAGGTCGTAATGTAAGGTAC................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................TGCCCGTGTATATATACCGATTTCTCTGT................................................................... | 29 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................TGTAGGTCGTAATGTAAGGTACCAAC............................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TATGTAGGTCGTAATGTAAGGTACCA............................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................CTGCATAAAACATTTAGCATAGCTTAGG.......................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................TGAAAACTGTTTGGCAAATGTGGC..................................................................................................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................TCTGTGCTGTGTACATATGTAGGTCGTA............................................ | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................TACATATGTAGGTCGTAATGTAAGGTACCA............................... | 30 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TTAGCATAGCTTAGGTTACTCTACA................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................AGGTCGTAATGTAAGGTACCAACAAC.......................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TATGTAGGTCGTAATGTAAGGTACCAAC............................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................TACCGATTTCTCTGTGCTGTGTACAT........................................................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................TTGGCAAATGTGGCTGCATAAAACATT........................................................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................ATACCGATTTCTCTGTGCTGT.............................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................TCTGCTCGTTATACCCCGCCCACGTG....................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................TGTGCTGTGTACATATGTAGGTCGTAA........................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................TACTCTACACCCTGCTCTAAGCATA................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................TGGCAAATGTGGCTGCATAAAACATT........................................................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................TGTACATATGTAGGTCGTAATGTA....................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................TGTAGGTCGTAATGTAAGGTACCAACAAC.......................... | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........TACTAAAACCATTTTCTGCTCGTTA...................................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................ACATATGTAGGTCGTAATGTAAGGTACC................................ | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................TCTGTGCTGTGTACATATGTAGGTCGT............................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................CAAGGTCGTAATGTAAGGTACC................................ | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................ACCGATTTCTCTGTGCTGTG............................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................TGAAAACTATTTGGCAAATGTGGCA.................................................................................................................................................................... | 25 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................TATATGGCAAATGTGG...................................................................................................................................................................... | 16 | 1 | 18 | 0.33 | 6 | 0 | 0 | 0 | 0 | 0 | 6 | 0 |
| ..............................................................................................................................................................................................................................ACAATAGCAGCAGCAATA.......... | 18 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................TATTTGGCTAATGTGGAT.................................................................................................................................................................... | 18 | 2 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................CTGTAAGGCACCAACAA........................... | 17 | 2 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CACCGTTTTAAATGATTTTGGTAAAAGACGAGCAATATGGGGCGGGTGCACTCGTTGATTTACTTTTGATAAACCGTTTACACCGACGTATTTTGTAAATCGTATCGAATCCAATGAGATGTGGGACGAGATTCGTATATATATATAACACACGACGGGCACATATATATGGCTAAAGAGACACGACACATGTATACATCCAGCATTACATTCCATGGTTGTTGTTGTCGTCGTCGTTATGTTGTTAGTT
*******************************************************........((((..(((((((....(((.....(((((...((....((((((((.......))))))))))..))))).....))).......))))))))))).....************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M053 female body |
M025 embryo |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
M023 head |
SRR618934 dsim w501 ovaries |
GSM343915 embryo |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................TTACTTTTTAGAAACCGTTTA.......................................................................................................................................................................... | 21 | 2 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................GAGCCAGATGGGGCGGGTG.......................................................................................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................GGGGGAAGAGATTCGTCTA............................................................................................................... | 19 | 3 | 16 | 0.25 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................TCCAATAAGATGTCTGACGA......................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................TAGGTATATATAACACAC................................................................................................. | 18 | 2 | 20 | 0.25 | 5 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 1 |
| ...........................................................TTACTTTTGTAGAACCGTTTAC......................................................................................................................................................................... | 22 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TTACTTTTTAGAAACCGTTT........................................................................................................................................................................... | 20 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................CGGGCTCATATATATGC.............................................................................. | 17 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GGGAAGAGATTCGTCTA............................................................................................................... | 17 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................TTGTTGTCGTCGTCGTT............ | 17 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................TTGATTTACGCTTGAAAAAC................................................................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................TCCAATTCGATGTGGGGC........................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................TGTTGTTGTCGTCGTCGT............. | 18 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................................GTTGTTGTCGTCGTCGT............. | 17 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 2r:12326861-12327110 - | dsi_13453 | G-TGGCAAAATTTACT-------AAAACCATTTTCT-GCTCGTT--ATA---CCCCG---------------------------CC----CAC-GTGAGCAAC-TAAATGA-AAACTATTTGGCAAATGTGGCTGCATAAAACA--TTTAGCA-TAGCTTAGGT-TACTCTACACCCTGC---------TCT----------------AAGCA---T---------------ATA-------------TATAT--ATTGTGTGCTGCCCGTGT----ATAT--ATA----CCGA------TTT------C------------------------------TCTGTGCTGTGTACA-T--------------ATGTAGGTCGTAATG------TA---AGGTACC--A----------------------------------------------------------------ACAACAACA---------GCAGCAGCAATACAAC-------AATCAA------ |
| droSec2 | scaffold_1:9154294-9154549 - | G-TGGCAAAATTTACT-------AAAACCATTTTCT-GCTCGTT--ATA---CCCCG---------------------------CC----CAC-GTCAGCAAC-TAAATGA-AAACTATTTGGCAAATGTGGCTGCATAAAACA--TTTAGCA-TAGCTTAGGT-TACTCTACACCCTGC---------TCT----------------AAGCA---TATATAT---------ATA-------------TATAT--ATTGTGTGCTGCCCGTGT----ATAT--ATA----CCGA------TTT------C------------------------------TCTGTGCTGTGTACA-T--------------ATGTAGGTCGTAATG------TA---AGGTACC--A----------------------------------------------------------------ACAACAACA---------GCAGCAGCAATACAAC-------AATCAA------ | |
| dm3 | chr2R:11660703-11660968 - | G-TGGCAAAATTTACT-------AAAACCATTTTCT-GCTCGTT--ATA---CCCCT---------------------------CC----CAC-GTGAGCAAC-TAAATGA-AAACTATTTGGCAAATGTGGCTGCATAAAACA--TTTAGCA-TAGCTTAGGT-TACTCTACACATTGC---------TCT----------------AAGCA---TATATAT--A---TAAATA-------------TGTAT--ATTGTCTGCTGCCCGTGT----ATAT--ATA----CCGA------TTT------C------------------------------TCTGTGCTGTGTACA-T--------------ATGTAGGTCGTAATG------TA---AGGTACC--A----------------------------------------------------------------ACAACAGCAACAACA---GCAGCAGCAATACAAC-------AATCAG------ | |
| droEre2 | scaffold_4845:14201977-14202203 + | C-TGGCGAAATTTACT-------AAAACCATTTTCT-GCTCGTT--ACA---CCCCG---------------------------CC----CTC-GTGAGCAAC-TAAATGA-AAACTATTTGGCAAATGTGGCTGCATAAAACA---TTAGCA-TAGCTTAGGT-TACTCTAC---CT---------------------------------------ATACTC---------GCA-------------CATAT--TTTGTCTGCTGCCCGTGT----ATAT--ACATATGCCCA------TTT------C------------------------------TCTGTGCCCTGTATG-T--------------ATGTAGGTCGTAATG------TA---AGGTACC--A----------------------------------------------------------------GCAGCAGCA----------------------------------------GCAG | |
| droYak3 | 2R:17326324-17326581 + | C-TGGCAAAATTTACT-------AAAACCATTTTCT-GCTCGTTTTATA---CCCCG-A-------------------------CC----CAC-GTGAGCAAC-TAAATGA-AAACTATTTGGCAAATGTGGCTGCATAAAACA--TTTAGCA-TAGCTTAGGT-TACTCTACA------------------------------------------TATA-TCGTAC--TATAGAACCTACATACCTATATAT--TTTGTCTGCTGCCCGTGT----ATAT--ATA----CCGA------TTT------C------------------------------TCTGTGCTGTGTACA-T--------------ATGTAGGTCGTAATG------TA---AGGTACC--A----------------------------------------------------------------ACAACAGC---------------AGCAGTACAAC-------AATCAG------ | |
| droEug1 | scf7180000409672:559696-559971 - | C-TGGCGAAATTTACT-------AAAACCATTTTCT-GCTCGTT--ATA---CCCCG---------------------------CCCCCACAC-GTGAGCAAC-TAAATGA-AAACTATTTGGCAAATGTGGCTGCATAAAACA--TTTAGCA-TAGTTTAAGT-TACTCTACACTC------AC----TCTACCTATCTATCTATCTAT-----CTATATAT---------ATA-------------TGTAT--ATTGTCTGCTGCCCGTGT----ATAT--ATA----CCGA------TTT------C------------------------------TCTGTGCTGTGTACA-T--------------ATGTAGGTCGTAATG------TA---AGGTACC--A----------------------------------------------------------------ACAGCAGC------------AACAGCAATACAAC-------AATCAGCAACAG | |
| droBia1 | scf7180000302292:3997758-3998018 + | C--GGCGAAATTTACT-------A-AACCATTTTCT-GCTCGTTC-TAA---CCCCG---------------------------CC----CCT-GTGAGCAAC-TCAATGA-AAACTATTTGGCAAATGTGGCTGCATAAAACG--TTTAGCA-TAGCTTAGGT-TACTCTACACCCCGCCATCTACAATCT----------------ATG------GT------ACACTTTAT---------------GTATTTCATGTCTGCTGCCCGTGT----ATAT--ATA----CCGA------TTT------C------------------------------TCTGTGCCGTGTACA-T--------------ATGTAGGTCGTAATG------TG---AGGTACC--A----------------------------------------------------------------GCAGCAG------------CAGCAGCAGTACAAC-------AATCAG------ | |
| droTak1 | scf7180000415241:19363-19635 + | C-TGGCGAAATTTACT-------A-AACCATTTTCT-GCTCGTTT-AAA--CCCCCG---------------------------CC----CAT-GTGAGCAAC-TAAATGA-AAACTATTTGGCAAATGTGGCTGCATAAAACG--TTTAGCA-TAGTTTAAGT-TACTCTACACTCT-----ATACACTCT----------------ATACACCCTATA------CACTATATA-------------TATGT--TTTGTCTGCTGCCCGTGT----ATAT--ATA----CCGA------TTT------C------------------------------TCTGTGCCGTGTACA-T--------------ATGTAGGTCGTAATG------TA---AGGTACC--A----------------------------------------------------------------ACAGCAGCAGCA------GCAACAGCAATACAAC-------AATCAG---CAA | |
| droEle1 | scf7180000491107:640270-640520 - | C-TGGCAAAATTTACT-------AAAACCATTTTCT-GCTCGTT--ATA---CCCCG---------------------------TT----CAC-GTGAGCAACTTAAATGA-AAACTATTTGGCAAATGTGGTTGCATAAAACA--TTTAGCA-TAGTTTAAGT-TACACTACACTCT---------------------------------------ATATAT---------ATA-------------TATAT--ATTGTCTGCTGCCTGTGTGT--ATAT--ATA----CCAA------TCT------T------------------------------TCTGTGCTGTGTACA-T--------------ATGTAGGTCGTAATG------TA---AGGTACC--A----------------------------------------------------------------ACAGCAGCAGCA------GCATCAGCAGTACAAC-------AATCAG------ | |
| droRho1 | scf7180000779911:451695-451928 + | T-TGGCGAAATTTACT-------A-AACCATTTTCT-GCTCGTT--AAA---CCCCG---------------------------TC----CAC-GTGAGCAAC-TAAATGA-AAACTATTTGGCAAATGTGGCTGCATAAAACA--TTTAGCA-TAGTTTAAGT-TACACT-----------------------------------------------------------TTATA-------------TATAT--ATTGTCTGCTGCTCGTGTGT--ATAT--ATA----CC-A------TTT------A------------------------------TCTGTGCTGTGTACA-T--------------ATGTAGGTCGTAATG------TA---AGGTACC--A----------------------------------------------------------------ACAGCAGCA---------GCAGCAGCAGTACAAC-------AATCAG------ | |
| droFic1 | scf7180000453809:155184-155457 - | C-TTGCGAAATTTACT-------A-AACCATTTTCT-GCTCGTTTTACA---CCCCG---------------------------CC----CAT-GTGAGCAAC-TAAATGA-AAACTATTTGGCAAATGTGGCTGCATAAAACA--CTCAGCA-TAGTTTAAGT-TACCCTACACAAC---------------------------------------AG------ACACCTTAGA-------------TATAT--AGTGTCTGCTGCCTCTGTA--TATAT--ATA----CCGATACCAATTCATCTGTG------------------------------TGTGTGCTGTGTACA-TAT------------ATGTAGGTCGTAATG------TA---AGGTACC--A----------------------------------------------------------------ACAGCAGCAGCAGCAGCAATATCAGCAGTACAAC-------AATCAG------ | |
| droKik1 | scf7180000302682:1835586-1835839 - | T-TGGCGAAATTTACT-------A-AACAATTTTCC-GCTCGTT--ATA---CCTCCCATG---CCCCTGCC-CCCTGTGATCGCCCC--CAC-GTGAGCAAC-TAAATGA-AAACTATTTGGCAACTGTGGCTGCATAAAGCA--TTTAGCACTAGACTA--T-TAG---ACA--CT-----AC----CCTACAT---------------TA---T---------------ATA-------------TATAT--ACTATATG--------------------------------------TT------C------------------------------TC-ATGCTGTGT-------------------ATGTAGGTCGTAATG------TA---AGGTACC--A----------------------------------------------------------------GCAGCAGCAGCAACAGCAGCAGCAGCAGTACAGT-------ACTCAG---CAG | |
| droAna3 | scaffold_13266:12539603-12539826 + | A-TGGCGAAATTTACT-------A-AACAATTCTCT--CTCGCT--C-----CCCCG------------------------------------------ACAC-TAAATGCAAAACTATTTGGCAAATGTGGCTGCAGAAAACA--CTTAGCA-TAGTTTAAGCCTCCTCTG----------------------------------------------------------------------------TATA-------------GACCGTCT----GTAT--ATA--------------TCT------C------------------------------TGTGTGCTGTGTACA-T--------------ATGTAGGCCGTAATG------TG---AGGTACC--CACCACA--------------G---CAACAACACCA-----------------------------CCAGCAGCAACA------GCAGCAGCAGTACAAC-------ACTCAG------ | |
| droBip1 | scf7180000396372:39489-39732 + | A-TGGCGAAATTTACT-------A-AACAATTCTCT-A--------------TCTTG---------------------------CT----CCC-CTG--AAAC-TGAATGCAAAACTATTTGGCAAATGTGGCTGCAGAAAACA--CTTAGCA-TAGTTTAAGC-C--TCCAC------------------------------------------------------------------------------------TGTCTATAGACCGTCTGTATGTAT--A--------TA------TCT------T------------------------------TGTGTGCTGTGTACA-T--------------ATGTAGGCCGTAATGTGAATGTG---AGGTACC--AGCCACA--------------G---CAACAGCAACA----C-------------------------CAGCAGCAGCAGCAGCAACAGCAGCAGTACAAC-------ACTCAG------ | |
| dp5 | 3:19524043-19524362 + | TTTGGCAAAATTTACTGAGAGCTAAAACAATTCTAT-G---GTT--ATATT-CCTCG-ACGGAACGCCTGAACCCCT--AAACAACTG--CATACTGTGCACG-TAAATGAGAAACTATTTGGCAAATGTGGCTGCAGCAAAGAATTTTGGCA-TAGTTTAAGT-TAC---ACACTCT-----AT----CCTGCATATATACATATAT---------------------------------------------------------AACTATAT----ATAT--ATA----TCTA------TCT----ATA------------------------------TCTATGCTGTATATA-TAT--ATATATACACATGTAGGTCGTAATG------TA---AGATACC--AACAACA--------------A------CAGC----AACACTAT------CAGCAACAACAATAC-CAACAACAACA---------------A------CAGCAGCTG----CAGCAG | |
| droPer2 | scaffold_37:526267-526603 + | TTTGGCAAAATTTACTGAGAGCTAAAACAATTCTAT-G---GTT--ATATT-CCTCG-ACGGAACGCCTGAACCCCT--GAACAACTG--CATACTGTGCACG-TAAATGAGAAACTATTTGGCAAATGTGGCTGCAGCAAAGAATTTTGGCA-TAGTTTAAGT-TAC---ACACTCT-----AT----CCTGCATATATACATATAT---------------------------------------------------------AACTATAT----ATAT--ATA----TCTA------TCT----ATA------------------------------TCTATGCTGTATATA-TATATATATATACACATGTAGGTCGTAATG------TA---ACATACC--AACAACAACAGA-AAC-CCCAA---CAACAGC----AACACTAT------CAGCAACAACAATAC-CAACAACAACA---------------A------CAGCTGCTG----CAGCAG | |
| droWil2 | scf2_1100000004512:926182-926385 + | --------------------------------------------------------------------------------------------------------TAAATGA-GAATCATTTGGCAGCT--TATTGCATA-----------------TCGTGAG-----------------------------------------AAA---------------------------------------------------------TTTCCATATGTATATAT--ATA----TCTA------T--------ACCTACCCAAAATACTACTACTACTATATCCCCCAAACTATGTATGTT--------------GTGTAGGTCGTAATG------TA---AGGTACC--AGCAACA--------------A---CAACAACAACA----CCAACATAATTAC-------------CAACAACAACA---------------ACAATATCAGCAGCAG----CAGCAG | |
| droVir3 | scaffold_12970:6946205-6946302 + | G-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCTATGT----ATGTATATA----T-AA------TAT------A------------------------------CATGTGCAATGCAAA-A--------------TTGGCAATTGCAA--------------------------------------------------GCGC----AAAGCTAT------CA-------------ACATTACCA---------GCATCAGCAATCACTT-------CATCAT------ | |
| droMoj3 | scaffold_6359:1876353-1876499 + | A-CACACAGATATA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTATATAT---------------------------------------------------------AAATATAT----ATGC--ACA----TATA------TAT----ATA------------------------------TAT----TTTAT-------------------TTGTATCTTGAAAT-------TT---GTGTGCC--AACAAAGAGAAC-GACGCCCGCCGGCAACAGCAGCA-----------------------------ACAGCAGCAACA---------GCAGCAAC------AGCAGCAAC----CGCAG | |
| droGri2 | scaffold_15245:5841156-5841375 - | C-CTGCTTAATTAACT-------T-AACCATCTAATTGCA-ATTG-AAA---C--------------------------------------------------------------------------TTTAAGTGTGTAAA-C--------AA-TAATTTAAGT-CG-------------------------------------------------------------------A-------------TGCAA--AAGTCCTTGAGTCTCTGTA--TATAT--ATA----TGAA------TAT------G------------------------------AAT--GG-TTATCTA-T--------------GTGTAGGTCGTAATG------TACAAAGGTACAACAGCCTCAACAGACAAC-AACAA---CAACAAC----AACACCAT------CA----------------GC---------------AACAACAATACCATCAGCAGCAG----CAGCAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/19/2015 at 05:07 AM