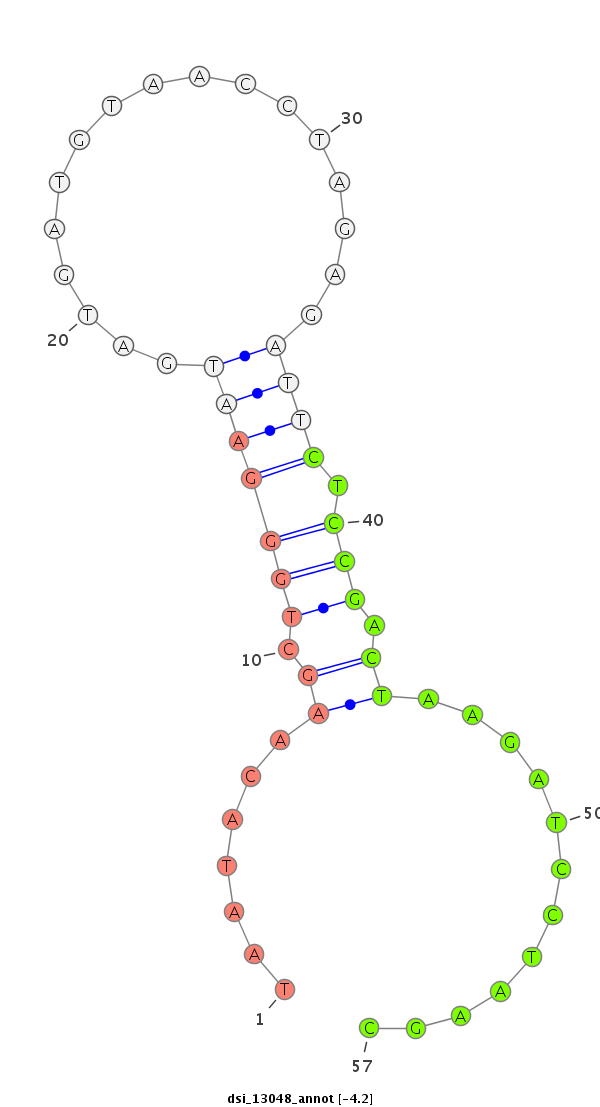
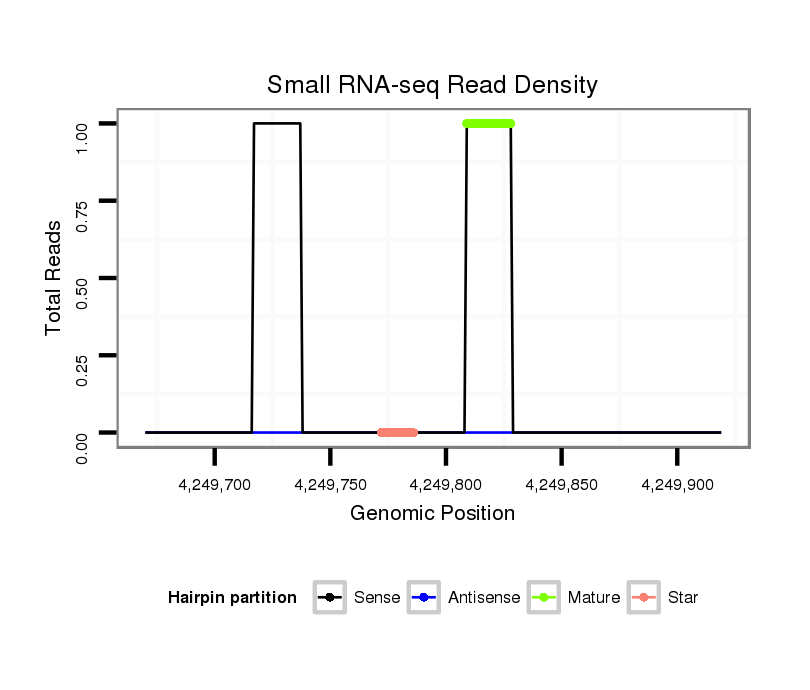
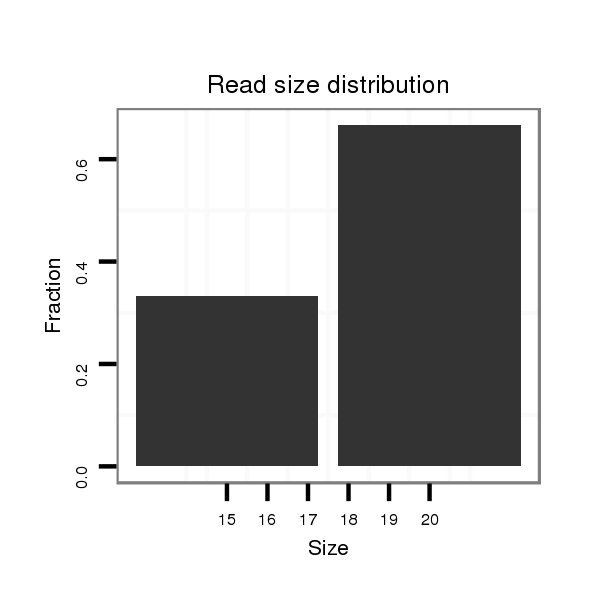
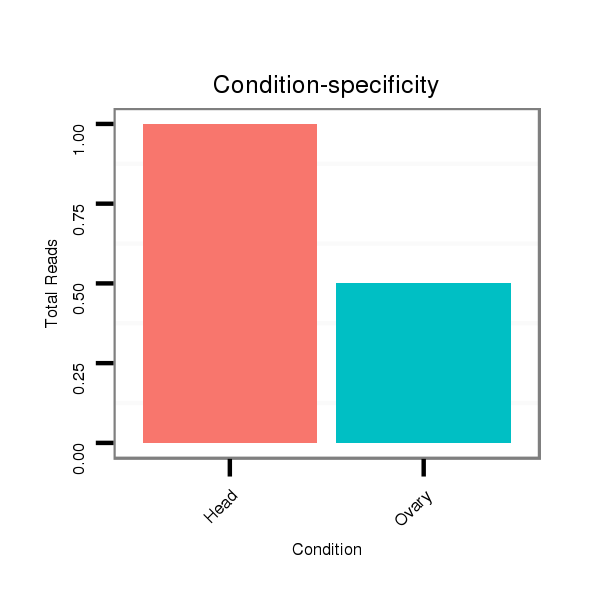
ID:dsi_13048 |
Coordinate:3r:4249720-4249869 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -4.6 | -4.2 | -4.2 |
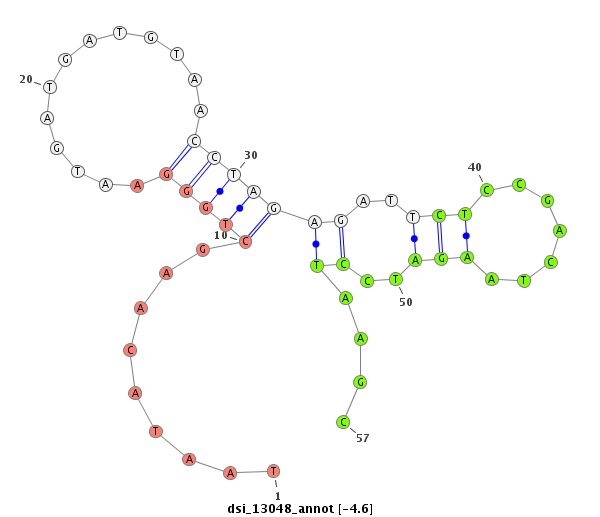 |
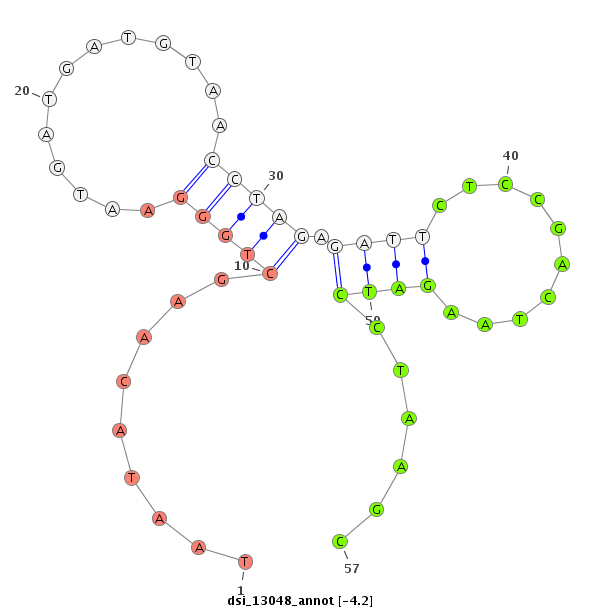 |
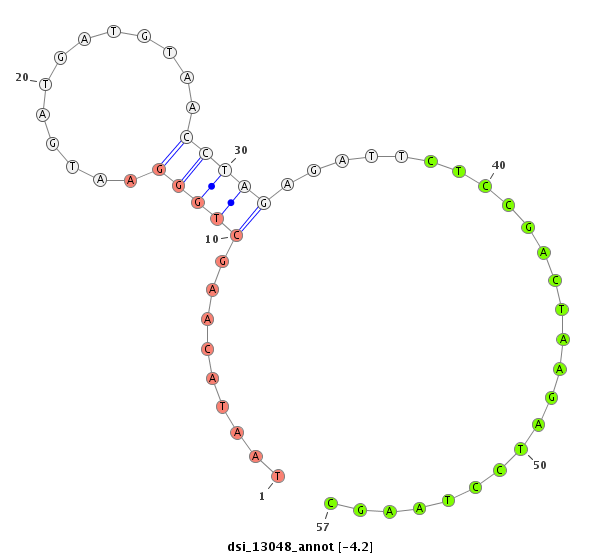 |
five_prime_UTR [3r_4249870_4249885_+]; CDS [3r_4249886_4250993_+]; exon [3r_4249870_4250993_+]; intron [3r_4245973_4249869_+]
No Repeatable elements found
| mature | star |
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TATAATAGTTTATTGTGCGAGCGCTCCATTAACACGAAGTGTTATAAATATGCATGTATGAATGAATTTAATTGCTGCCGCAAGGCATTAACCTGCATTGCATAATACAAGCTGGGAATGATGATGTAACCTAGAGATTCTCCGACTAAGATCCTAAGCCCAAAGCTGCCTAAATACAACTAACAAAGTATTCCCCACAGTTAAAATATCTTCAAGATGGGTTCGCTGCCACAATTGTCGATCGTCAAGG ******************************************************************************************************.......((.(((((((.................)))).))).))............******************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
M023 head |
SRR902008 ovaries |
M025 embryo |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................ATATGCATGTATGAATGAATT...................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................CTCCGACTAAGATCCTAAGC........................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................TAATACAAGCTGGGA..................................................................................................................................... | 15 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TCTGGTACTGATGATGTAAC........................................................................................................................ | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................ATACTACCAGCTGGGACTGA................................................................................................................................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................TAACATCCTAAACCCAATGCT................................................................................... | 21 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................ATACGAGCTGGGAATG.................................................................................................................................. | 16 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 |
|
ATATTATCAAATAACACGCTCGCGAGGTAATTGTGCTTCACAATATTTATACGTACATACTTACTTAAATTAACGACGGCGTTCCGTAATTGGACGTAACGTATTATGTTCGACCCTTACTACTACATTGGATCTCTAAGAGGCTGATTCTAGGATTCGGGTTTCGACGGATTTATGTTGATTGTTTCATAAGGGGTGTCAATTTTATAGAAGTTCTACCCAAGCGACGGTGTTAACAGCTAGCAGTTCC
*******************************************************************************************.......((.(((((((.................)))).))).))............****************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM343915 embryo |
M023 head |
M024 male body |
|---|---|---|---|---|---|---|---|---|
| ...................................................CGTACATACTTACGGAAATTAT................................................................................................................................................................................. | 22 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ...................TCGCAAGGTGATTGAGCTTC................................................................................................................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 |
| .....................................................................................................................................................................................................................................GTAGTAACAGCTTGCAGTT.. | 19 | 3 | 20 | 0.10 | 2 | 0 | 2 | 0 |
| ...............................................................................................................................................................................TGTCGATTGTTGCACAAGG........................................................ | 19 | 3 | 17 | 0.06 | 1 | 0 | 1 | 0 |
| ......................CGAGGTCATTGTTCTTGAC................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
Generated: 04/23/2015 at 11:26 PM