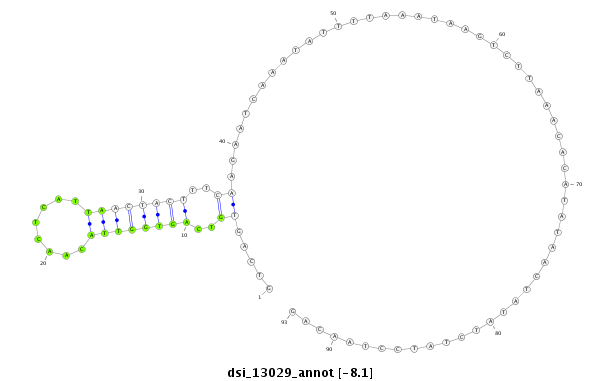
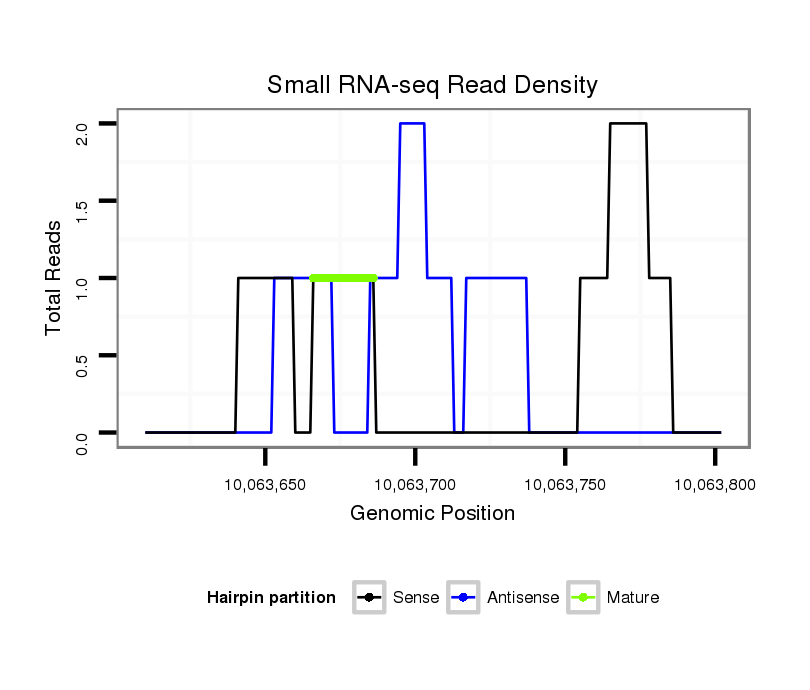
ID:dsi_13029 |
Coordinate:3r:10063660-10063752 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
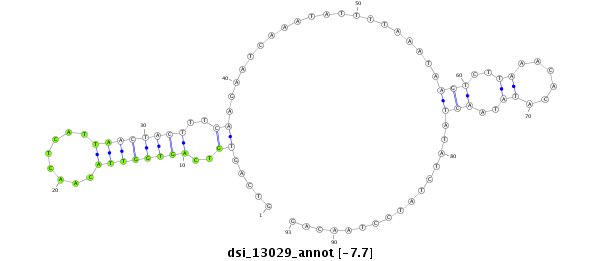
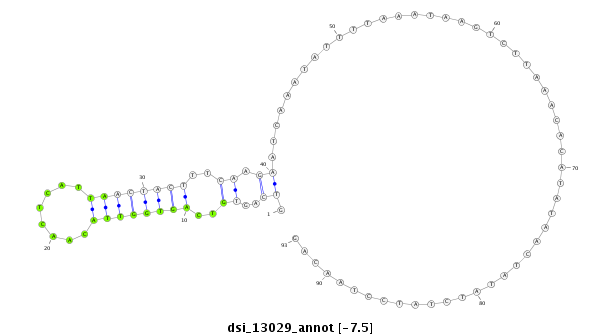
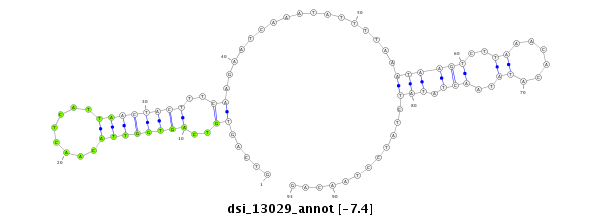
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -7.7 | -7.5 | -7.4 |
 |
 |
 |
exon [3r_10063449_10063659_+]; CDS [3r_10063449_10063659_+]; CDS [3r_10063753_10063978_+]; exon [3r_10063753_10063978_+]; intron [3r_10063660_10063752_+]
No Repeatable elements found
| ##################################################---------------------------------------------------------------------------------------------################################################## AGGAGACGCCCCTCACAAGTCTGGGATTAAGCCTCAGCGGCTGCGAGCAGGTCAGTGTCAGTGGTTACAACTCATTAACTACTTTCAAGAATCAAATATTTTAAATAAGTCTTAAACACATATAACTATATCTATCCTAACAGGAGGCCACCAATCGCGAGCTGGAGCAGGAGCTGCGCACCACGTTTCCGGA **************************************************.....((..((((((((........))))))))..))........................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
SRR618934 dsim w501 ovaries |
M023 head |
O002 Head |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................CGCGAGCTGGAGCAGGAGCTG................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................CCTCAGCGGCTGCGAGCAG............................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................GGCCACCAATCGCGAGCCGGAGC......................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................GGCCACCAATCGCGAGCTGGAGC......................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................GTCAGTGGTTACAACTCATTA.................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................TCTGGAATTAAGCCT............................................................................................................................................................... | 15 | 1 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 1 | 1 |
| ...............CGATTCTGGAATTAAGCCT............................................................................................................................................................... | 19 | 3 | 12 | 0.33 | 4 | 0 | 0 | 0 | 0 | 4 | 0 |
| .................................................................................................................................................GGCCACCAATCCCGAGC............................... | 17 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................ATTCTGGAATTAAGCCT............................................................................................................................................................... | 17 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
|
TCCTCTGCGGGGAGTGTTCAGACCCTAATTCGGAGTCGCCGACGCTCGTCCAGTCACAGTCACCAATGTTGAGTAATTGATGAAAGTTCTTAGTTTATAAAATTTATTCAGAATTTGTGTATATTGATATAGATAGGATTGTCCTCCGGTGGTTAGCGCTCGACCTCGTCCTCGACGCGTGGTGCAAAGGCCT
**************************************************.....((..((((((((........))))))))..))........................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
O002 Head |
M024 male body |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
M053 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................GTTCTTAGTTTATAAAAT.......................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................ATTGATGAAAGTTCTTAGT................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................GCTCGTCCAGTCACAGTCAC.................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCAGAATTTGTGTATATTGAT................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................GCCAGTCACAGTCACCAATGT............................................................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................TGTTGAGTAATAGATG............................................................................................................... | 16 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......CGGGGAGAGTTCAGA........................................................................................................................................................................... | 15 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................TCAACAATGTTGAGGAAT.................................................................................................................... | 18 | 2 | 6 | 0.33 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................TCGCAGTCACCAATTGTG.......................................................................................................................... | 18 | 3 | 20 | 0.10 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................AGATAGGACTGTCCTCG.............................................. | 17 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................CGGAGTGGCCCACGCT................................................................................................................................................... | 16 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................TCACAGTCAGCAATGAAG.......................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
Generated: 04/24/2015 at 10:24 AM