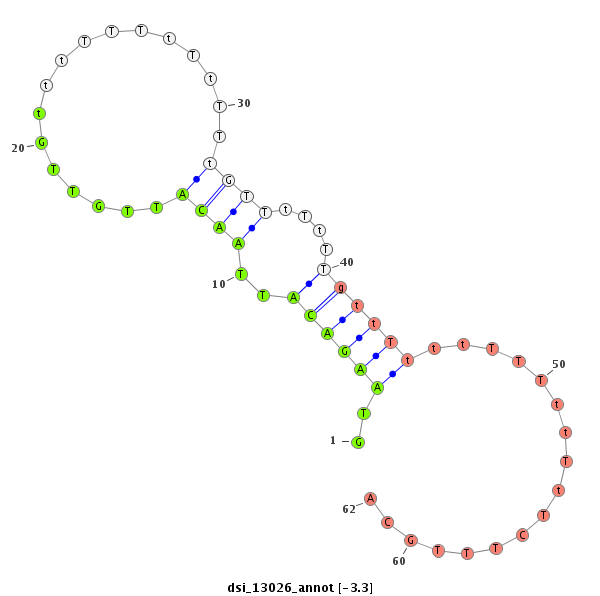
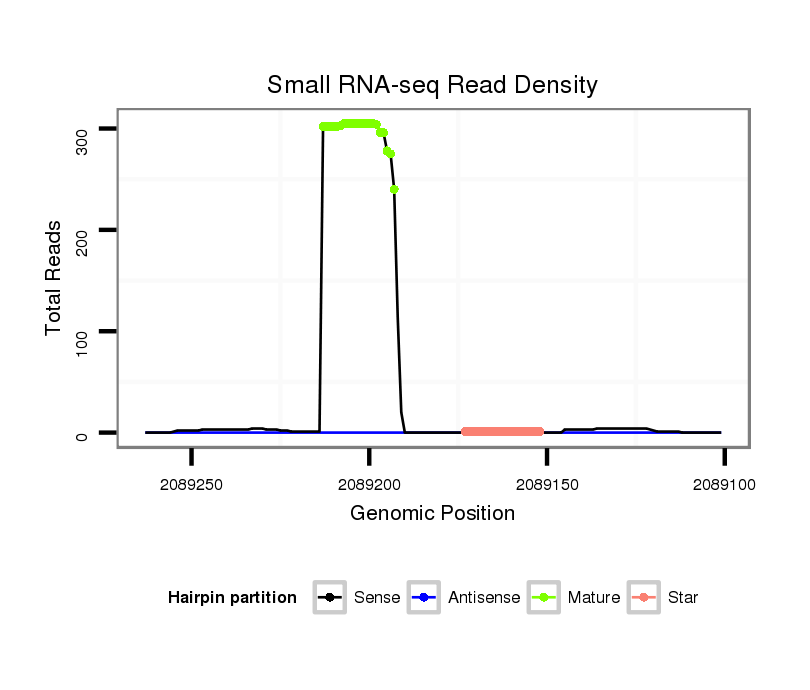

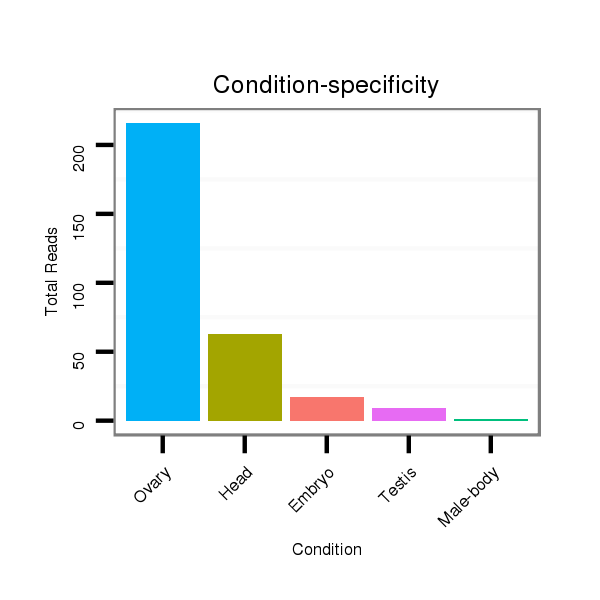
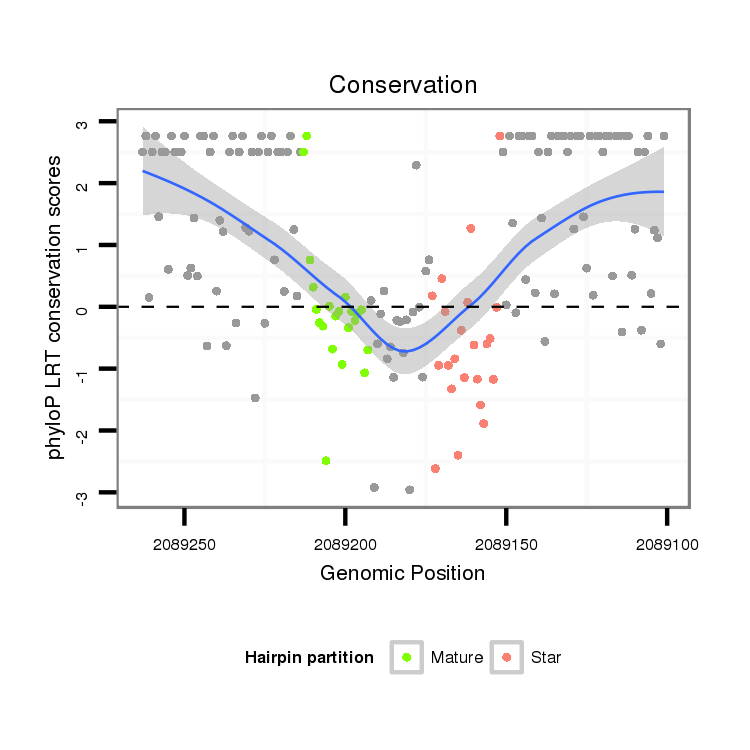
ID:dsi_13026 |
Coordinate:3r:2089151-2089213 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -2.6 | -2.4 |
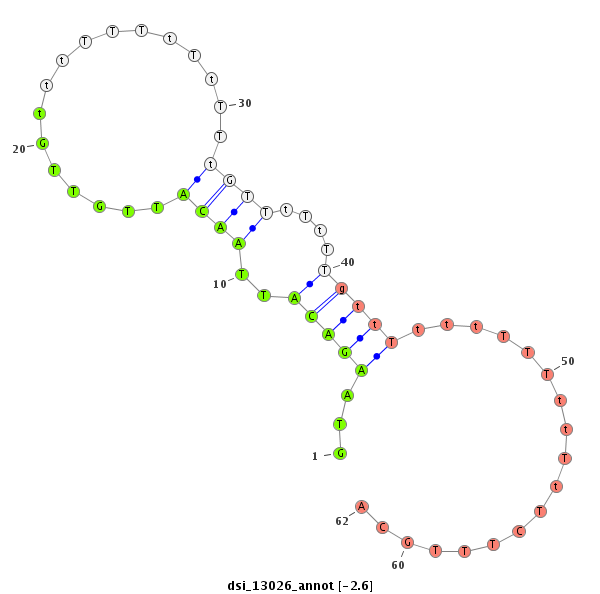 |
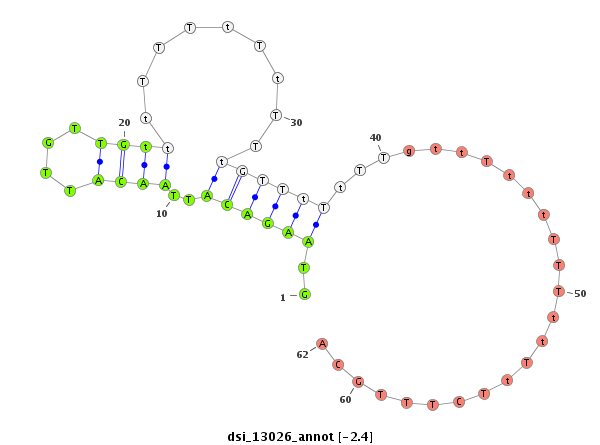 |
exon [3r_2088821_2089150_-]; exon [3r_2089214_2089578_-]; CDS [3r_2088821_2089150_-]; CDS [3r_2089214_2089578_-]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ##################################################---------------------------------------------------------------################################################## GATCATCGTTGGGAACTAATCGAAAGCCTTGACCGTGAGGATCGCGAGCGGTAAGACATTAACATTGTTGtttTTTtTtTTtGTTtTtTTgttTtttTTTttTtTCTTTGCAGAAAATTCAACGAGCACATTGACAATCTGATGAAGAAGAAGAGGGAGCGGT **************************************************..((((((..((((.................))))....)))))).................*************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
M023 head |
O002 Head |
M025 embryo |
O001 Testis |
GSM343915 embryo |
SRR902009 testis |
M024 male body |
SRR553486 Makindu_3 day-old ovaries |
M053 female body |
SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGACATTAACATTGTTGA............................................................................................ | 21 | 0 | 1 | 126.00 | 126 | 86 | 10 | 22 | 4 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGACATTAACATTGTTGAA........................................................................................... | 22 | 0 | 1 | 92.00 | 92 | 72 | 1 | 2 | 9 | 5 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGACATTAACATTGTTG............................................................................................. | 20 | 0 | 1 | 35.00 | 35 | 14 | 18 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGACATTAACATTGTTGAAA.......................................................................................... | 23 | 0 | 1 | 19.00 | 19 | 18 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGACATTAACATTGT............................................................................................... | 18 | 0 | 1 | 18.00 | 18 | 12 | 1 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGACATTAACATT................................................................................................. | 16 | 0 | 1 | 8.00 | 8 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGACATTAACATTGTTGG............................................................................................ | 21 | 1 | 1 | 5.00 | 5 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGACATTAACATTGTT.............................................................................................. | 19 | 0 | 1 | 3.00 | 3 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGACATTAACATTGTGG............................................................................................. | 20 | 2 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGACATTAACATTGTTGAAAAA........................................................................................ | 25 | 2 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................CATTAACATTGTTGAA........................................................................................... | 16 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................ACATTAACATTGTTGAA........................................................................................... | 17 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................TCAACGAGCACATTGACAATCTGAT.................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGACATTAACATTGTTGAG........................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................TCAACGAGCACATTGACAATCTGATG................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGACATTAACAT.................................................................................................. | 15 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................GACCGTGAGGATCGCGAGCG................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................CATTAACATTGTTGAAA.......................................................................................... | 17 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................ACATTGACAATCTGATGAAGAAGA............ | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................GTAAGACATTAACATTGA............................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........TGGGAACTAATCGAAAGCCTTGACCGTGA............................................................................................................................. | 29 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................CAATAAATTTAATATCTTTGCA................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................TCAACGAGCACATTGACAATCTGA..................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGACATTAACATTGTTGCA........................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................TAATCGAAAGCCTTGACCGTGAGGA.......................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................GTAAGACATTAACATTTGA.............................................................................................. | 19 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........TTGGGAACTAATCGAAAGCCTTGACC................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGACATTAACATTGTTGT............................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGACATTAACATTGTTGTA........................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................GAAGAAGAAGAGGGA..... | 15 | 0 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CTAGTAGCAACCCTTGATTAGCTTTCGGAACTGGCACTCCTAGCGCTCGCCATTCTGTAATTGTAACAACtttAAAtAtAAtCAAtAtAAgttAtttAAAttAtAGAAACGTCTTTTAAGTTGCTCGTGTAACTGTTAGACTACTTCTTCTTCTCCCTCGCCA
***************************************************..((((((..((((.................))))....)))))).................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|
| ......................................................................................................................TCTTGCTCGTGTAAC.............................. | 15 | 2 | 6 | 0.17 | 1 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3r:2089101-2089263 - | dsi_13026 | GATCATCGTTGGGAACTAATCGAAAGCCTTGACCGTGAGGATCGCGAGCGGTAAGACA--TT-A------ACA-----TTGTTGtt--------tTT------Tt-------------T--------tTTtGTTt---T-tTT-gttTtttTT--Ttt------------------------------TtTCTTTGCAGAAAA-TTCAACGAGCACATTGACAATCTGATGAAGAAGAAGAGGGAGCGGT |
| droSec2 | scaffold_6:2273116-2273279 - | GATCATCGCTGGGAACTAATCGAAAGCCTTGACCGCGAGGATCGCGAGCGGTAAGACA--TT-A------ACA-----TTGTTGAA--------ATT------TA-------------T--------ATTTAGTT--AT-ATT-CAATAAATT--TAA------------------------------TATCGTTGCAGAAAATTTCAACGAGCACATTGACAATCTGATGAAGAAGAAGAG-GAGCGGT | |
| dm3 | chr3R:2188007-2188170 - | GATCATCGCTGGGAACTAATAGAAAGCCTTGATCGCGAGGATCGCGAGCGGTAAGGCA--TT-A------ACA-----TTGTTAAA--------AAT------TC-------------T--------ATTTATTT--AT-ATT-CAATAAATT--TAA------------------------------TATCGTTGCAGAAAA-TTCAACGAGCACATTGACAATCTGATGAAGAAGAAGAGGGAGCGAT | |
| droEre2 | scaffold_4770:2432546-2432708 - | GACCATCGCTGGGAACTAATTGAAAGCCTCGACCGCGAGGATCGCGAGCGGTAATACA--TT-A------ACA-----TTACTAAA--------TAT------AA-------------A--------ATTTAGTTTT----A-AATAAAAATG--TAA------------------------------TACCATTACAGAAAA-TTCAACGAGCACATTGACAATCTGATGAAGAAGAAGCGGGAGCGAT | |
| droYak3 | 3R:18113527-18113691 - | GACCATCGCTGGGAACTAATAGAAAGCCTCGACCGCGAGGATCGCGAGCGGTAACACA--TT-A------ACA-----TTACTTAA--------AAT------TA-------------A--------AGTAAAAT--AA-ATTAAAATAAATA--TAA------------------------------TACAATTACAGAAAA-TTCAACGAGCACATTGACAATCTGATGAAGAAGAAGCGGGAGCGAT | |
| droEug1 | scf7180000409504:18545-18700 - | GACCATCGCTGGGAACTGATCGAGACTCTTGACCGAGAAGATCGCGAGAGGTAAGAAT--CT-AT-----ACA-----ATT---------ATTT---------------AAAT------A----------------------------------------------AT--CAAT-TAATAATAATTTG-ACTTGCGCAGAAAA-TTCAATGAACATATTGATAATCTGATGAAGAAAAAGCGAGAGCGAT | |
| droBia1 | scf7180000302155:2515479-2515640 + | GACCACCGCTGGGAACTGATCGAGACCCTCGATCGAGAAGATCGCGAGCGGTAAACAA---CA-T-----TGA-C-A-T-----------ATTCATT------A------------------------------------------------------GCAAGTCGATCTTAGTA---TATAAACTTT-TCTTTTACAGGAAG-TTCAACGAGCACATTGACAATCTGATGAAGAAGAAGCGGGAGCGAT | |
| droTak1 | scf7180000415261:469789-469954 + | GACCATCGCTGGGAACTGATCGAGACTCTCGATCGGGAAGATCGTGAGCGGTAAAAATAAACA-T-----ATA-----CTT---------ATTC--A------TA-------------------------------------------------A--AGCATGTTAATCTTATTA---TATTAACTTC-TTGTCAACAGAAAG-TTCAACGAGCACATTGACAATCTGATGAAGAAAAAGCGAGAGCGAT | |
| droEle1 | scf7180000491278:925617-925776 - | GACCATCGCTGGGAACTGATTGAGACCCTCGATCGAGAGGATCGCGAGCGGTAGGGAA--AA-AA-----ACA-----TTTTTC---------T---------------GAAT------A---------TTT--T-T---------------------------------CAGCATACAACTAACTTT-ACGCTTACAGCAAA-TTCAACGAGCACATTGACAATTTAATGAAAAAGAAGCGGGAGCGAT | |
| droRho1 | scf7180000779776:50600-50759 + | GACCATCGCTGGGAACTGATTGAGACCCTCGATCGAGAAGACCGCGAGCGGTAAGAGG--AC-A------ACA-----AATTAT-A--------TAA---------------T------A---------TTT--T----------------------AGT----------GTAT-TACGAATAACTTC-ACCTTTACAGCAAA-TTCAACGAGCACATTGACAACCTGATGAAAAAAAAGCGAGAGCGGT | |
| droFic1 | scf7180000453910:545959-546115 - | GATCATCGCTGGGAACTGATCGAGACTCTCGATCGAGAAGATCGCGAGCGGTAAGAAG--AC-AA-----AAA-----TTT---------ATTT---------------AA-T------A---------ACT--T---------A---------T--A----------------ATACGATAAAATTT-ACTTTCATAGAAAA-TTCAACGAACACATTGACAATCTGATGAAGAAAAAGCGAGAACGTT | |
| droKik1 | scf7180000302639:622828-623000 + | GATCATCGCTGGGAGCTGATCGAAACACTCGATAGGGACGATCGTGAGCGGTAAGGAT--AT-AAATTAAAAA--ATATA-----------------------------------ATATAATTTATATAATATTT--ATA-T-AAATTATCCCCTGC--------------------------------AATAATTCAGGAAA-TTCAACGAGCACATTGACAATCTAATGAAGAAAAAGCGCGAACGGT | |
| droAna3 | scaffold_13340:13423390-13423555 - | GATCATCGCTGGGAGCTGATAGAGACTCTTGATCGGGATGATCGTGAGCGGTTAGTAA--TA-G------GCT-----TTTTTTGA--------AAA------TCAT------------TCT-----TTT----TTTAA-ATT-ACATAATAT--TAC------------------------------TATCCCTACAGAAAG-TTCAATGAACATATTGACAATCTCATGAAGAAAAAGCGCGAGCGCT | |
| droBip1 | scf7180000396691:2287310-2287469 - | GACCATCGCTGGGAGCTGATAGAGACTCTTGATCGGGACGATCGTGAGCGGTTAGTAG--TA-CA-----CCA-----TTTTTTAA--------CA----------------T------ACT-----GTT----TTT---------------------------------AAACGGAAAAGTAC----TTTCACTTTAGAAAA-TTCAACGAGCATATTGACAATCTCATGAAGAAGAAGCGCGAGCGCT | |
| dp5 | 2:20204973-20205132 + | GATCATCGATGGGAGCAGATCGAGCCACTCGATCGAGATGATCGTGAGAGGTAGGTTCAGAA------------------------ATCCA---------------TTACA-TACATA------------------------------------T--AGT----------AAATACCATAGTTACTTT-CATCTTGTAGAATC-TTCAATGTACACATTGACAATCTGATGAAGAAGAAACGCGAGAGAT | |
| droPer2 | scaffold_3:2949882-2950041 + | GATCATCGATGGGAGCAGATCGAGCCACTCGATCGGGATGATCGTGAGAGGTAGGTTCAGAA------------------------ATCCA---------------TTACA-TACATA------------------------------------T--AGT----------AAATACCATAGTTACTTT-CATCTTGTAGAATC-TTCAATGTACACATTGACAATCTGATGAAGAAGAAACGCGAGAGAT | |
| droWil2 | scf2_1100000004902:10613791-10613953 - | GATCATCGCTGGGAACTGATCGAAACGCTAGATCGGGATGATCGCGAGAGGTATATATAGAA---------TATT-A-AAT---G------T--ACA------TGAA------------ACT-----ATA----TCT---------------------------------AATCT---CTTAATCACT-CCTTCGATAGAATT-TTTAACGAACACATTGACAATCTGATGAAAAAGAAACGAGAGAAAT | |
| droVir3 | scaffold_13047:17345847-17346011 - | GATCATCGCTGGGAACTAATTGAGACACTCGATCGTGATGAACGTGAGCGGTAAGTAA--ACTA------GCG-----ATAATTG------------TAAGATT---------------------AGCACCTGAT--ATT-T-GATGTCATTG--T-------------------------------A-TTTCTTCCAGTATC-TTTAACGAACATATTGACAATTTGATGAAAAAGAAGCGCGAAAAAT | |
| droMoj3 | scaffold_6540:14313200-14313364 + | GATCATCGCTGGGAATTAATCGAGACACTTGATCGTGATGATCGTGAACGGTGAGCCT--ACG-T-----GGA-----ACA-----AGTTA------TTAAATT---------------------AGCATATTTT--ATT-T-CA----------------------------------ATTATTGTC-TATCGCGCAGAATC-TTCAACGAGCATATTGACAATCTGATGAAAAAGAAACGCGAAAAAT | |
| droGri2 | scaffold_14906:12403500-12403672 - | GATCATCGCTGGGAATTGATTGAGACACTCGATCGCGATGATCGTGAGCGGTAAGTCT--TC-CATCCCAGCG--ACATTCTGT--------------TAAATT---------------------GGCATTTTTT--ATC-T-TAAATCTCGT--GTA------------------------------TCCGTTCGCAGAATC-TTTAACGAGCATATTGACAATCTGATGAAAAAGAAACGCGAAAAGT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 10/12/2014 at 02:47 PM