| droSim2 |
x:6283757-6284135 - |
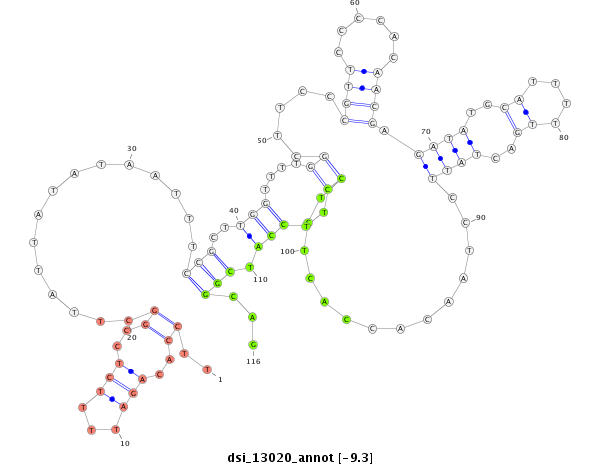
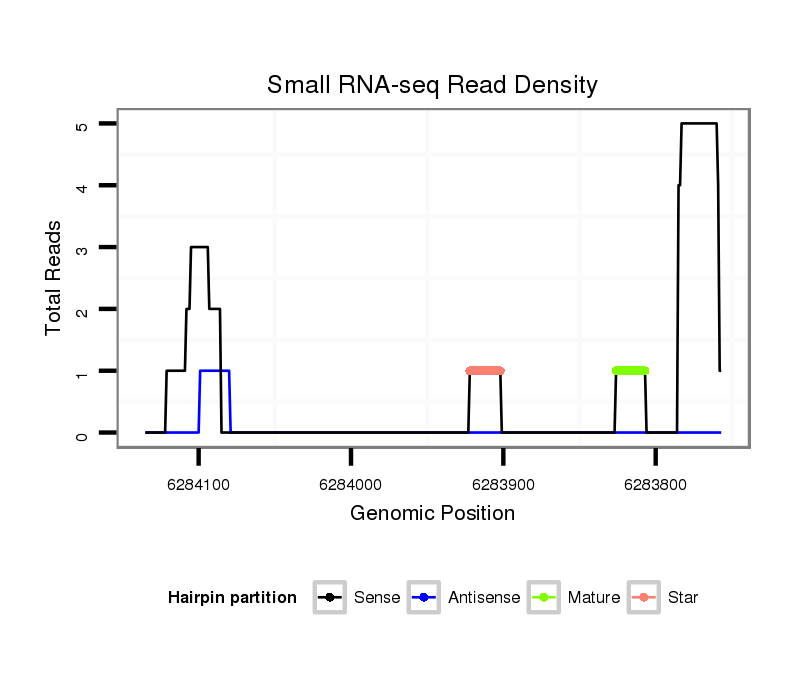
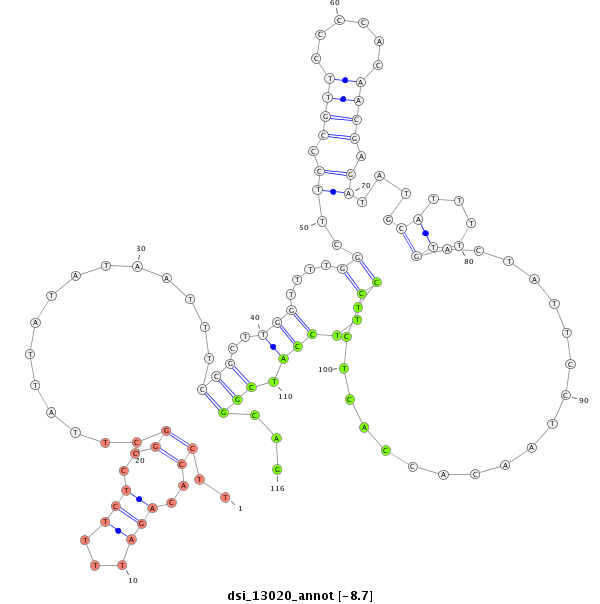
dsi_13020 |
CGTCTGG---TGGCAGGGAAA--------------------------------CT-AACAGCAATCGCCTCTCAAACGAATCGGGTACCCAGAACA----------TCTATCA-----------T-C-------GTACTTAAAATCCCGCTT-----ATTAT-TTTGTATCTCTTGCACG---------CGCCCAATGA-ATCCCCTCCC-----------------------------------------A-A----------------------------------------------------------------------------------------------------------------------------GTTTTCGTTAGGCGTA---TTGTATACATGTTTGTCC-------------TAGATGCT---GTAAATAGTT-GGCTTTATCCTC--CCGC--------TTCGCCCA---------GA-----TCCTTTCCA---CAGATTTTCT-CCG------------GCTTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATATAATTTCCGCTTGGTTTT------GGCTTCCCGTTCCCCAC--------------------------------------------------------------------------------------------------AACGAGA------TATGCATTTT-TGA---CTATTCCTAACAC-CACTCT------CCTTCC-ATCGGCAGGTTTCGTGTCGATGCGTGAGTTTCGCACTTGCACACAGAC---------------GGCGGACTAC |
| droSec2 |
scaffold_4:114150-114528 + |
|
CGTCTGG---TGGCAGGGAAA--------------------------------CT-AACAGCAATCGCCTCTCAAACGAATCGGGTACCCAGAACA----------TCTATCA-----------T-C-------GTACTTAAAATCCCGCTT-----ATTAT-TTTGTATCTGTTGCACG---------CGCCCAATGA-ATCCCCTCCC-----------------------------------------A-A----------------------------------------------------------------------------------------------------------------------------TTTTTCGTTAGGCGTG---TTGTACACATGTTTGTCC-------------TAGATGCT---GTAAATAGTT-GGCTTTATCCTC--CCGC--------TTCGCCCT---------GA-----TCCTTTCCA---CAGATTTTCT-CCG------------GCTTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------TTGTATAATTTCCGCTTGGTTTT------GGCTTCCCGTTCCCCAC--------------------------------------------------------------------------------------------------AACGAGA------TATGCATTTT-TGA---CTATTGCTAACAC-CACTCT------CCTTCC-ATCGGCAGGTTTCGTGTCGATGCGTGAGTTTCGCACTTGCACACAGTC---------------GGCGGACTAC |
| dm3 |
chrX:6680796-6681176 - |
|
CGTCTGG---TGACGGGGAAA--------------------------------CT-AACAGCAATCGCCACTCAAACGAATCGGGTACCCAGAACA----------TCTATCA-----------T-C-------GTACTTAAAATCCCGCTT-----ATTAT-TTTGTATCTGTTGCACG---------CGCCCAATGA-ATTCCCTTCC-----------------------------------------AAA----------------------------------------------------------------------------------------------------------------------------TTTTTCGTTAGGCGTA---TTGTATACATGTTTGTCC-------------TAGATGCT---GTAAATAGTT-GGCTTTATTCTC--CCGCC-------TCCGTAAT---------GA-----TCCTTTCCA---CAGATTTCCT-CCG------------GCTTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATATAATTTCCGCTTGGTTTT------GGCTACCCGTTCCCCAC--------------------------------------------------------------------------------------------------AACGAGA------ACTGCATTTT-TGA---CCATTTCTAACAC-CACTCT------CCTTCC-ATCGCCAGGTTTCGTGTCGATGCGTGAGTTTCGCACTTCCACACAGAC---------------GACGGACTAC |
| droEre2 |
scaffold_4690:15608714-15609089 - |
|
CGACTGG---TGGCGGGGAAA--------------------------------CT-AACAGCAATCGCCTCTCAAACGAATCGGGTACCCAGAACA----------TCAGTCA-----------T-C-------CTACTTAAAATCCCGCTT-----CTGAT-TTTGTAACTGTTGCACG---------TGCCCAATGA-ATCCCCTCCC-----------------------------------------A-A----------------------------------------------------------------------------------------------------------------------------TTTATCGTTAGGCTTA--ATTGTACATATGTTTTGCC-------------T--TTGTA---GTAAATAGTT-GGCTTTATCCCT--CCGCC-------CGCCTCCT---------GA-----TCCGTTCCC---CAGTTTGTCT-CCG------------GCATA---------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATTTAATTTGCGCTTGGTTTT------GGCTACCTGTTTCCCAC--------------------------------------------------------------------------------------------------AGCGAGT------TCCTTATTTT-TGA---CTATTTGTAACAA----TCT------CCTTCC-ACCGGCAGGTTTCGTGTCGATGCGCGAGTTTCGCACTTCCACACAGAC---------------GACGGACTAC |
| droYak3 |
X:2646712-2647109 - |
|
CGACTGG---TGGCGGGGAAA--------------------------------CT-AACAGCAATCGCCACTCAAACGAATCGGGTACCCAGAACA----------TCTATCA-----------T-C-------GTACTTAAAATCCCGCTT-----ATTAT-TTTGTATCTGTTGCACGGCCAATGCACGGCCAATGA-ATCCCCTCCC-----------------------------------------A-A----------------------------------------------------------------------------------------------------------------------------TTCATCGTTAGGCATT---TTGTATCTATGTTTGTCCAGA--CTCATCAATAGATGCT---GTAAAAAGTT-GGCTTTATCCTC--CCGCC-------TTCCACCT---------GA-----TCCTTTCCA---CGGCTTTCCT-TCG------------GCTTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATTTAATTATCGCTTGGTTTT------GGCTTCCAATTACCCAC--------------------------------------------------------------------------------------------------AACGAGG------TCTGCACTTTCGGA---CTATTTCTAACGC----TCT------CCTTCC-ATCGGCAGGTTTCGTGTCGATGCGCGAGTTTCGCACTTCCACACAGAC---------------GACGGACTAC |
| droEug1 |
scf7180000409095:7353-7810 - |
|
CGTCTAG---TGGCGGGGAAA--------------------------------CT-AACAGCAATCGCCACTCAAACGAATCGGGTACCCAGAACA----------TCTATAA-----------T-C-------GTAGTTACAGTCCCGCTT-----AATAGATTTATAT--------------------------------------------ATATATATATAAATCGTATATTCCCTTGCCCTTCCC-G-ACTTTCGTTACTCCTTTTTTGATCTCATGGCTTAATTTTAGTTTTTA--------AAGTATACGCATTATAT------------------TAAAGCTTTTCCAT----------------------TTCCCATTAGGCTTTTC-CTGTATATATTTTT----AAGCACATATCGTTTAAGGCT---GTAAATAGTTTGGCTTGATTCCC--CCAC--------CTTTCCTT---------TT-----TCCTTTCCA---CTGCTTTTCT-CCT------------GTATA---------------------------------------------------------------------------------------------------------------------------------------------------------------------TTGTTTAATTTCCGCTTGGTTTT------GGCATTCTGTTCCCCAT---------------------------------------------------------------------------------------------------------------------TTAT-GGA---CTATTTTTAACCA----TCT------CCTTCC-ATCGGCAGGTTTCGTTTCGATGCGCGAGTTCCGCACCTCCACGCAGAC---------------GACGGACTAC |
| droBia1 |
scf7180000301760:1925877-1926342 + |
|
CGGCTGG---TGGCGGGGAGA--------------------------------CT-AACAGCAATCGCCACTCAAACGAATCGGGTACCCAGAACA----------TCTATCA-----------T-C-------GTACTTACAATCCCGCTTTACGTATAAT-TTTGTATCCGTTGCACC---------TGGCGAAATA-CACCCCTCCGATCCA-------CTTAATG-------------------------------------CGC-TTTG---TAAAGACGCCATTTTATTTTGTAAGGCTTT-----ATAC-CATTAGA--AAATAAGTTTGAGCACTCAGAGCTTTGGCCT----------------------------TTGGGCCTT---CTGTATATACTTC-C--CACA--CACATCGATAGCTGCT---GTAAATAGTTTGACCT------C--CAAC--------CTTTTCTTTATACCGCCTATCCTGCCCTTCCCA---C---TTTCCC-TGT------------GTATA---------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCTTTAATTTCCGCTTGGTTTTGGATTTGGCTTCCCATTCCCCGC--------------------------------------------------------------------------------------------------AACGTAT------GCTAAGTTAT-GGC---CTATTTTTAAACC--TCTCT-CTCCACCTTCC-CCCGGCAGGTTTCGTCTCGATGCGCGAGTTCCAG--------------------------------------- |
| droTak1 |
scf7180000413955:145269-145406 - |
|
TCTCTC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGT------------GTATA---------------------------------------------------------------------------------------------------------------------------------------------------------------------CTGCTTAATTTCCGCTTGGTTTT------GGCTCCCTATTCCCCCC--------------------------------------------------------------------------------------------------GTCGCCT----------CCTTTT-AGAACTCTATTTTTAACCC--AGTCTCCTCCTCCTTCCAACCGACAGGCTTCGTTTCGATGCGCGAGTTCCAG--------------------------------------- |
| droEle1 |
scf7180000491044:595079-595559 - |
|
TGGCTGG---TGGCGGGGAAA--------------------------------CT-AACAGCAATCGCCACTCAAACGAATCGGGTACCCAAAAA----CAAATAATCTATCA-----------A-C-------GTAGTTACAAGCCTGCTT-----TATAT-TTTGTATCCGTTATCCA----------ACCTGGTGACATTTCCTTTCCTGCATAAATATGTTAATG------------------------------------------TTG---TAA-AGCTTAATTG-------GCAGACTTTAAAAT---TGCATAAAAATAAATACATTGGAG--------GCTTTAGCCTTT--------------------CTCACACTAGGCTTT---CTGTACATATATTTT--TAAA--CACATCAACAGCTGGTGTAA-AAATAGTTTGGCTTGATTCCCTTCCC----------------------------------CCCTCCCA---CTGCTTCTCT-CCTCTAT----ATT-ATTTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATTTACTTTTCGCTTGGTTTT------GGCTTGCTGTTTACC-T----------------------------------------------------------------------------------GTTACCTGTTATCCACAACTCAC------ACTGATTTAT-GGA---CTATTTTTAACGC----TCT------CCTTTCGACCGGCAGGTTTCGTTTCGATGCGTGAGTTTCGCACCTCCACGCAAAC---------------AG-------- |
| droRho1 |
scf7180000776942:275281-275539 - |
|
TTC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACGCTAGGCTTT---CTGTATATATTTTT----AAA--CACATCGACAAATGCT---GTAAATAGTTTGACTTCATTCCT-CCCGCCCTCCCGCCTTTTCCC---------TTTC---TTCTCCCTA---CTGCTTTTCT-CCT------------GTGTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATTTAATTTCCGCTTGGTTTT------GGCTTCCTATTCCCCAC--------------------------------------------------------------------------------------------------AACGTAC------TCTGAATTAT-GGA---CTATTTTTAACAC----TCT------CCTTCC-GCCGGCAGGTTTCGTTTCGATGCGCGAGTTTCGCACCTCCACGCAGAC---------------GACGGA---- |
| droFic1 |
scf7180000454072:727864-728321 + |
|
CGACTGGTGGTGGCGGGGAGA--------------------------------CT-AACAGCAATCGCCACTCAAACGAATCGGGTACCCAGAAAA-AACAAACAA-CTAT------C-TACTATCC-------GTAGCTACCATCCCGACC-----TGCAGCTCC-T---------------------------------------C------------------------------CTCCTCCTCCTCTA-CTTTCCACTTCCCTCCC--------------------------------------------------------------------G--------GCTTTCTTCCTCCCCATGCTCCATGTGCTCCTCCATGCTTTGGCTTG---CTGTACACCTGTTCC--CGAG--CAGGTCGACAGATGCT---GTAAATAGTTCGGCTTGGCTCCT--CC--------------TCTC---------CTCC---GCCTTCCCA---CCGCCG-TCC-CCT------------TTAAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------CTGTTTAATTTCCGCTTGGTTTC------GGCTTCCTGCTCCCCGTTTTCTCTTCCCTCTTCTCTGTTATCCGTTCTCTTTTATCCGTTCATTGTTATCCGTTCTCTGTTATCCGTTCTCTGTTATATGTTAACTGTTCTC---CACGAGAACACACTCTCCAGCAC-GGG---TTCTTTTTAA------------------------------------------------------------------------------------------------ |
| droKik1 |
scf7180000302610:385216-385588 - |
|
GG--------CGGCAGCAGCA--------------------------------CTCACCAGCAATCGGCACTCCAATGAATCCGGTACCTACTCACAGACAGAAAATCCT--A---GCTCACTAGCT-------GT-TACAAAGTTCTGCTC-----CTCCT-GC-----------------------------TATGA-------------------------------------------------------GTTCCGTTACCCCTCC--------------------------------------------------------------------G--------CCCTCCC------------------------CCTTTCCCTAGTTATC---CTGGACCTGCGCCTGT----------------ATATAGC---ATAAATGC--------------CA-CT--C-------CTCCTCCT---------GC-----TTTCTCCCC---TCGTT----------TAT----TTTTG-TTTTTGTATTCCCCCTTTTTGGTTATGGCTTTCTTGTTGATTTTTTTCTGCCTTTTCTTGGCTTTTCTCTT------------------------------ATTTTTGTTGTTGTTATTGTTG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGT-TCCCTTAGGTTTTGTGTCCATGAAGTCGTTTCGCACCTCCACGCAGAG---------------CGTGTCCAAG |
| droAna3 |
scaffold_13117:4471083-4471452 - |
|
TGGCTGG---TGGCGGGGAAAGTAATACGGAACTGCGGAAAGCAGCCGCGGCACTCACCATCAATCGCCATTCCAACGAATCGGGTACTGGCT--A----------TGACCTATGACC-TATGACCTATGACTAGCACTACTACTTCCGCTA-----CTA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCACTACTAGTCTGCTTCTAT----TTCTGCTTCTTGTAATGCCCGCTT---------------------------------------------------TAAGTGTTGT--AGTCTACTCCTCTTCCCTTTAATTTTGTTACCATT--TTTTGTGTCCACATCCGCTTAATCCTCAAGAGCTTTCAAGAGCTTCT---------------------------------------------------------------------------------------------------------------------------------------------------AA----------------------------------CTGACTC-TGCTCC------CTCTCC-TGCTTTAGGTTTCGTGTCCATGAAGTCATATCGCACCTCCACCCAAAGCGTGTCGAAGCGCTCCTCGGACTAC |
| droBip1 |
scf7180000395928:29979-30291 + |
|
CGCAAGG---CGGCCGCGGCA--------------------------------CTCACCATCAACCGCCATTCGAACGAATCGGGTACT--GGCTA----------TGACCTA-----------T-T-------GTACTACCACTTCCGCTA-----CTA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCACTACTAG----CTTCTACTAGCTCCTGCTTTCTGTAATATCCGC-------------------------------------TTGCTTGGTTTTTCTTTGAGTGTTGTGTAGCC-----TTCGCTTCCTTAGTTTTGTTCCC------------TCAGTGTCTCCTCAACCCACCTCAGCCCA-------CTCTAATT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------GGA---T------------------TCCTTCTCCT-CC-CGGTACAGGTTTCGTGTCCATGAAGTCGTTCCGCACCTCCACCCAGAGCGTGTCCAAGCGCTCCTCGGACTAC |
| dp5 |
XL_group1a:6505864-6505974 - |
|
TTCCTAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTCTATTCCC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTCTTTTT--CGTCCA----------CTCTTC-TGCGGCAGGTTTCGTGTCGATGAAGTCGTTCCGCACATCCACGCAGAGCGTCTCCAAGCGCTCCTCGGACTAC |
| droPer2 |
scaffold_25:809414-809524 + |
|
TTCCTAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTCTATTCCC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTCTCTTT--CGTCCA----------CTCTTC-TGCGGCAGGTTTCGTGTCGATGAAGTCGTTCCGCACATCCACGCAGAGCGTCTCCAAGCGCTCCTCGGACTAC |
| droWil2 |
scf2_1100000004909:7980945-7980997 + |
|
CGTAAGG---CAGCCGATATT--------------------------------CTAACCAGCAATCGTCATTCAAATGAATCGGGTAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droVir3 |
scaffold_12928:4045953-4046014 - |
|
CGC--------------------------------------------------TA-ACCAGCAATCGTCATTCGAATGAATCGGGTATTCCAGGCA----------ATTACTT-----------A-T-------ATATCTAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droMoj3 |
scaffold_6359:635337-635427 - |
|
CGCAAAG---CGGCGGCTGAG--------------------------------TTAACCAGCAATCGTCATTCAAATGAATCGGGTATTTCTTCTA------------------------------------------------------TA-----CTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTACT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCTTAGCAACTTCTATCGCT----------- |
| droGri2 |
scaffold_14853:621704-621772 + |
|
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAGGTTTCGTGTCGATGATGTCATTTCGTACGTCCTCGCATAGCGTTTCCAAGCGATCCTCTGAGCAC |