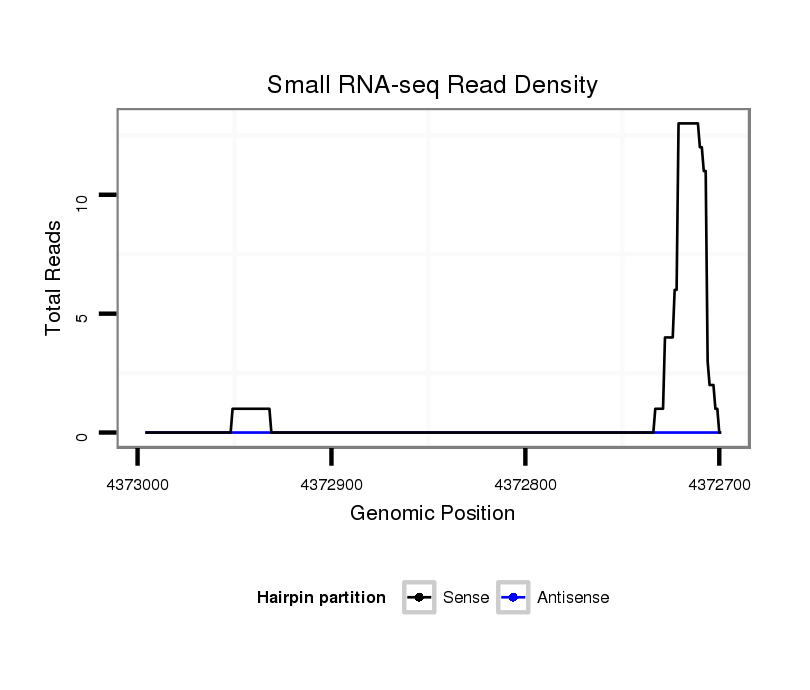
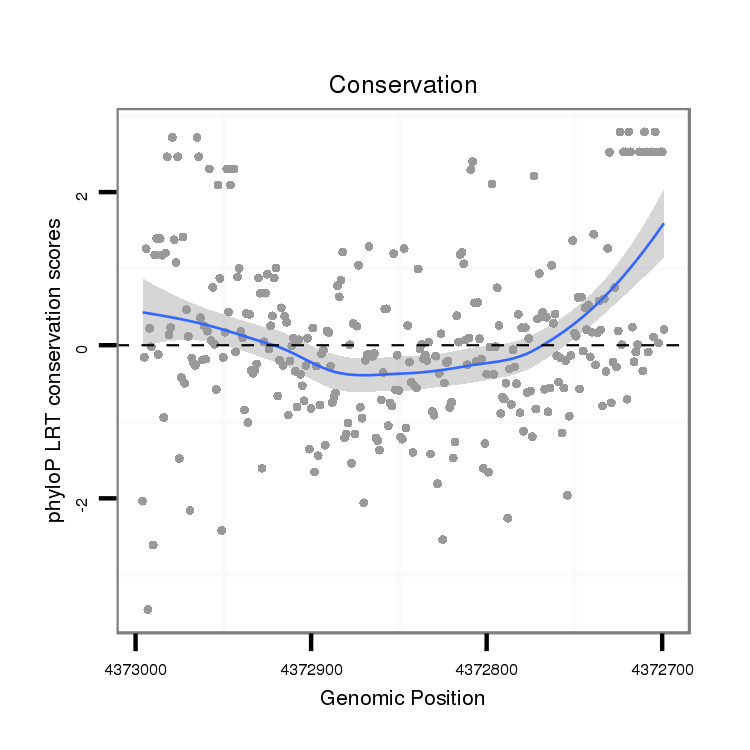
ID:dsi_12871 |
Coordinate:2l:4372749-4372946 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
intergenic
No Repeatable elements found
|
ACCCCACTTCACAGGCAATCCACTGCGTGCATGCGTCGAGAGGCCTTCAAGTATGACTTTGTCCTGGGATGGGCCGACTGGTTTGGAAAACCAATCATATACAACTCGGATTACTTAAAATGTTGCCTAAATTAAAGCTTTTCGATCTTATTCTTTGATGTCAGATTTGTCATCCAAACAAATCGGCCCAACTCCGTATACACCCTATAGATCAGAAGTATTGACCTAACTCCTACTTCCTCACATAGATATGTACCTACCCTTGCCCAAAGACCCTTGTAGACCGTCGCCTTGTGGC
**************************************************.((((....(((....(((((((((((...(((((((..((.((((..((((((...((((...((.(((.(((...........))).))).))....)))).))).)))..)))).))..)))))))...))))))))..)))....)))............((((((..((......)).))))))....)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
M023 head |
M025 embryo |
M053 female body |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
O001 Testis |
O002 Head |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR1275483 Male prepupae |
SRR1275487 Male larvae |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCGCT....... | 18 | 2 | 1 | 827.00 | 827 | 410 | 319 | 51 | 10 | 4 | 8 | 7 | 8 | 4 | 3 | 2 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCGCTT...... | 19 | 2 | 1 | 434.00 | 434 | 243 | 143 | 17 | 20 | 0 | 3 | 0 | 1 | 5 | 1 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCGCC....... | 18 | 1 | 1 | 120.00 | 120 | 59 | 57 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................CTTGTAGACCGTCGCTTGCT... | 20 | 3 | 5 | 99.80 | 499 | 309 | 136 | 26 | 12 | 4 | 2 | 0 | 4 | 0 | 5 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................................................................................................................................................TCTTGTAGACCGTCGCTT...... | 18 | 2 | 2 | 39.50 | 79 | 39 | 35 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCGCTTG..... | 20 | 3 | 5 | 21.60 | 108 | 60 | 12 | 5 | 19 | 1 | 2 | 0 | 3 | 3 | 0 | 1 | 0 | 1 | 1 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCGC........ | 17 | 1 | 1 | 14.00 | 14 | 0 | 0 | 0 | 0 | 10 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................CTTGTAGACCGTCGCTTG..... | 18 | 2 | 4 | 11.25 | 45 | 29 | 13 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCGCG....... | 18 | 2 | 2 | 9.00 | 18 | 9 | 7 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................TCTCTTGTAGACCGTCGCT....... | 19 | 3 | 6 | 8.67 | 52 | 18 | 24 | 2 | 1 | 0 | 2 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................TTGTAGACCGTCGCTTGCT... | 19 | 3 | 19 | 7.53 | 143 | 69 | 52 | 8 | 3 | 2 | 4 | 1 | 0 | 0 | 2 | 1 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................CTTGTAGACCGTCGC........ | 15 | 0 | 1 | 7.00 | 7 | 0 | 0 | 0 | 0 | 5 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................TCTCTTGTAGACCGTCGC........ | 18 | 2 | 2 | 6.50 | 13 | 6 | 5 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................TCTTGTAGACCGTCGC........ | 16 | 1 | 2 | 6.00 | 12 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CCCTTGTAGACCGTCGCTTGCT... | 22 | 3 | 1 | 5.00 | 5 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................TCTCTTGTAGACCGTCGCC....... | 19 | 2 | 1 | 5.00 | 5 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................TCTCTTGTAGACCGTCGCTT...... | 20 | 3 | 3 | 4.67 | 14 | 7 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................GTCTCTTGTAGACCGTCGCT....... | 20 | 3 | 4 | 4.50 | 18 | 7 | 7 | 0 | 1 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................GTCTCTTGTAGACCGTCGCTT...... | 21 | 3 | 2 | 3.00 | 6 | 1 | 2 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCGCTTTCT... | 22 | 3 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................CTTGTAGACCGTCGCT....... | 16 | 1 | 1 | 3.00 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................CTTGTAGACCGTCGCCTGCT... | 20 | 2 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCG......... | 16 | 1 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTATACCGTCGCC....... | 18 | 2 | 2 | 2.00 | 4 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................TCTTGTAGACCGTCGCT....... | 17 | 2 | 6 | 1.50 | 9 | 0 | 0 | 0 | 0 | 0 | 3 | 4 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTC.......... | 15 | 1 | 3 | 1.33 | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................TGTAGACCGTCGCTT...... | 15 | 1 | 3 | 1.33 | 4 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCGCTC...... | 19 | 3 | 7 | 1.29 | 9 | 4 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................ATCCTTTGTTGTCTGATTTGT................................................................................................................................ | 21 | 3 | 7 | 1.14 | 8 | 5 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CCCTTGTAGACCGTCGCTT...... | 19 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................AAAGACCCTTGTAGACCG............ | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CCCTTGTAGACCGTCGCTTGC.... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................TTGTAGACCGTCGCTT...... | 16 | 1 | 2 | 1.00 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................TCTTGTAGACCGTCGCCT...... | 18 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CCCTTGTAGACCGTCGCT....... | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................CTTGTAGACCGTCGCTTTCT... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................AAAGACCCTTGTAGACCGTCGCCTTG.... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCGCCTGCT... | 22 | 3 | 2 | 1.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTATACCGTCGCTT...... | 19 | 3 | 7 | 1.00 | 7 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................TTCAAGTATGACTTTGTCCT......................................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CCCTTGTAGACCGTCGCC....... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CCCTTGTAGACCGTCGCTTG..... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCGCTTGGT... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CCCTTGTAGACCGTCGCCTTGTG.. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCGCTTGG.... | 21 | 3 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................AAAGACCCTTGTAGACCGTC.......... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................TGCCCAAAGACCCTTGTAGACCGTCGC........ | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CCCTTGTATACCGTCGCT....... | 18 | 2 | 3 | 0.67 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCTTCGCTT...... | 19 | 3 | 11 | 0.64 | 7 | 5 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCGCTG...... | 19 | 3 | 5 | 0.60 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCGCGTGG.... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................CCTTGTAGACCGTCGCTTGCT... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................AGGTCTCTTGTAGACCGTCGC........ | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTAGCC....... | 18 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................ACCCTATAGATCCGAATTGTT............................................................................ | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCTTCGCC....... | 18 | 2 | 4 | 0.50 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCTTCGCT....... | 18 | 3 | 20 | 0.45 | 9 | 2 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................GGGATGGGCCGGCCGGT........................................................................................................................................................................................................................ | 17 | 2 | 7 | 0.43 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................CTTGTAGACCGTCGCTTAC.... | 19 | 3 | 7 | 0.43 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................TTGTAGACCGTCGCT....... | 15 | 1 | 5 | 0.40 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................TCTCTTGTAGACCGTCGCG....... | 19 | 3 | 10 | 0.40 | 4 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................ACCCTATAGATCCGAA................................................................................. | 16 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCGCCTGC.... | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................GCTCTTGTAGACCGTCGCTT...... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................AGTCTCTTGTAGACCGTCGCT....... | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CACTTGTAGACCGTCGCT....... | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTACACCGTCGCC....... | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCCCTT...... | 19 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................AGGTCTCTTGTAGACCGTC.......... | 19 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................GTAGACCGTCGCTTTCT... | 17 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................TCTTGTAGACCGTCG......... | 15 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................AGTCTTTGTTGTCTGATTTGT................................................................................................................................ | 21 | 3 | 10 | 0.20 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................TTGTAGACCGTCGCTTGCTG.. | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................TCTTTGTTGTCTGATTTGT................................................................................................................................ | 19 | 2 | 12 | 0.17 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCTCC....... | 18 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGCAGACCGTCGCC....... | 18 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................CCTAGTAGAACGTCGC........ | 16 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................CCTCTTGTAGACCGTCGCT....... | 19 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGTCACTT...... | 19 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................TTGTAGACCGTCGCTTG..... | 17 | 2 | 18 | 0.11 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCCTCGCTT...... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCCTCGCT....... | 18 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGCCGCTT...... | 19 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................ATCTTGTAGACCGTCGCTT...... | 19 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTCGTAGACCGTCGCT....... | 18 | 3 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTATACCGTCGCG....... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................ATGCCTTTGTCCTGG....................................................................................................................................................................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................TGTAGACCGTCGCTTG..... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................GTCTTTGTTGTCTGATTTGT................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................GCCTCGTAGACCGTCG......... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................TTCTTTGATGTCAAA..................................................................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................TCTTTTAGACCGTCGCTT...... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................TCTCTTGTAGACCCTCGC........ | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................CCTAGTAGAACGTCGCG....... | 17 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................TGCTTGCGTCGAGAGA............................................................................................................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CTCTTGTAGACCGCCGCT....... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................TCTCTTGTAGACCGTC.......... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
TGGGGTGAAGTGTCCGTTAGGTGACGCACGTACGCAGCTCTCCGGAAGTTCATACTGAAACAGGACCCTACCCGGCTGACCAAACCTTTTGGTTAGTATATGTTGAGCCTAATGAATTTTACAACGGATTTAATTTCGAAAAGCTAGAATAAGAAACTACAGTCTAAACAGTAGGTTTGTTTAGCCGGGTTGAGGCATATGTGGGATATCTAGTCTTCATAACTGGATTGAGGATGAAGGAGTGTATCTATACATGGATGGGAACGGGTTTCTGGGAACATCTGGCAGCGGAACACCG
**************************************************.((((....(((....(((((((((((...(((((((..((.((((..((((((...((((...((.(((.(((...........))).))).))....)))).))).)))..)))).))..)))))))...))))))))..)))....)))............((((((..((......)).))))))....)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM343915 embryo |
SRR618934 dsim w501 ovaries |
M025 embryo |
M024 male body |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................ATATCTAAAAGCTAGAATAA.................................................................................................................................................. | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......GAAGTCTCGGTTAGGTGCC................................................................................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.25 | 5 | 0 | 0 | 3 | 2 | 0 | 0 | 0 | 0 |
| ...................................................GTACTGAAACAGGACC....................................................................................................................................................................................................................................... | 16 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................TATGGGAACTGGCTTCTGG....................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................ATGGGAACTGGCTTCTG........................ | 17 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................AGTAGGTTTGTTCAG.................................................................................................................. | 15 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................................................................ATGAAGGAGGGTAAGTATA.............................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 2l:4372699-4372996 - | dsi_12871 | ACCCCACTTCACAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAATCCACTGCGTGCATGCGTCGAGAGGCCTTCAAGTATGACTTTGTCCT------------GGGATGGGCCGACTGGTTTGGAAAACCAAT------C-----ATATACAACTC-----------------------GGATTACTT---AA--AATGT---------------------------------------------TGCCTA----------------------AATTAAA-------------GCTT---------TTCGATCTTATTCT--------TTGATG---T--CAGATT--------------------------------------------TGT-------------------------------------------CATCCAAACAAATCGGCCCAACTCCGTATACACCCTAT-AGA--TCAGAAGTATTGACCTAACTCCT----A--CT--TCCTCA----------------------------------CATAGATATGTACCTACCCTTGCCCAAAGACCCTTGTAGACCGTCGCCTTGTGGC--------------------- |
| droSec2 | scaffold_5:2623888-2624174 - | ACCCCACTTCACAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAATCCACTGCGCGCATGCGTCGAGAGGCCTTCAAGTATGACTATGTCCT------------GGGGTGGGCCGACTTGTTTGGAAAAC----------------------AACTC-----------------------GGATTACTT---AA--AATTT---------------------------------------------TGCCTA----------------------AATTAAA-------------GCTT---------TTCGATCTTATTCT--------TTGATG---T--CAGATT--------------------------------------------TGT-------------------------------------------CATCCAAACAAATCGGCCCAACTCGTTATACACCCTAT-GGA--TCAGAAGTATTGAGCTAACTCCT----A--CC--TCCTAA----------------------------------AATAGATATGTACCTACCCTTGCCCAAAGACCCTTGTAGACCGTCGCCTTGTGGC--------------------- | |
| dm3 | chr2L:4525355-4525653 - | ACCCCACTTAACAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAATCCACTGCGCGCATGCGTCGAGAGGCCCTCAAGTATGACTTTGTCCT------------GGGATGGGTCGACTTGTTTGGAAAACCAAT------C-----ACATACAACTC-----------------------GCATTACTT---AA--ACTTT---------------------------------------------TTCCTA----------------------ACTTAAA-------------ACTT---------ATCGATCGTATTCT--------TTGATG---T--CAGATT--------------------------------------------TGT-------------------------------------------CATCCAAACAAATCGGCCCAACTCGTTATACACCATAT-AGA--TCAGAAGTATTGACCTAACTCCTA---A--CT--TATTCA----------------------------------CATAGATATGTACCTACCCCTGCCCAAAGACCCTTGTAGACCGTCGCCTTGTGGC--------------------- | |
| droEre2 | scaffold_4929:4615655-4615949 - | ACCTCACTTCACAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAATCCCCTGCGCGCATGCGTCGAGAGGCCCTCAAGTATGACTTTGTCCT------------GGGCTGGGTCGATTTGTTTGGAAAACGAAT------CGTTTTATCTTAAACTC-----------------------GGATTACTC---AGCTAATTT---------------------------------------------TGTCTA----------------------AATTATG-------------ACTT----------TGGTTCT--------------------------TAGCTTTGGT---TC---T--------------------AC----------TT-------------------------------------------CACCCAAAGAAATCGACCCAACTCGTTATACACCTTA-------TCAGGAGTCTTGAACTTAATGTT----A--CT--TCCCCT----------------------------------TATAGACATGTACCTTCCCCTGCCCAAAGACCCTTGTAGACCGTCGCCTTGTGGC--------------------- | |
| droYak3 | 2L:4553180-4553483 - | ACCTCACTTCACAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAATCCACTGCGCGCATGCGTCGAGAGGCCCACAAGTATGACTTTGTCCT------------GGGTTGGGTCGACTTGTTTGGAGAACGAAT------CCCAATGTTTTAGACTC-----------------------GGATTACTC---AACTAATTT---------------------------------------------TGCATG----------------------AATTTTA-------------GCTT----------TGGTTCTTATTTT--------TTGATG---A--CAGATT--------------------------------------------TGT-------------------------------------------CATCCAAACAAATCGACCCAGCTCGTTGTACACCTTAT-AGA--TCAGGAGTCTTTACCTAAATGCT----A--CT--TCCCCT----------------------------------TATAGACATGTACCTTCCCCTGCCCAAAGACCCTTGTAGACCGTCGCCTTGTGGC--------------------- | |
| droEug1 | scf7180000409005:410881-411225 - | ACCAGATTTCACCG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAATCCGCTGCGAGCATGCGTCAAGAGACCTTCAAGTATGACTTTGTCCT------------GGGGTGGGTCGACTTGAAGAGAACACGAAT------TG-----TAGTTACTTTATAATCTTTATTCTCTAAAAGTTGGT------------------TTTA----------------------------AGGCTGAAATAATTTCTTAAAATAGGAAAAACTGAACTGTAATCTATGAGTTCAGATC----------------------------TC--------GTT---CGTCTTCTCTGG---A-----------------------------------------TT-------------------------------------------CCACCAATCAAATCGACCCACCTCTCAG--CAACCTTT-AGA--CCAGAATTCTTGATCTAACTGTTTTGC-CC-T--CCTCCT----------------------------------CATAGATATGTACCTGCCCCTGCCAAAAGACCCTTGTAGACCCTCTCCTTGTGGC--------------------- | |
| droBia1 | scf7180000302188:1085790-1086112 - | GCCGGGTTTCACCG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAATCCACTGCGCGCATGCGTCGAAAGGCCCTCAAGTATGACTCTATCCT------------GGGGTGGGTCGATTTGTGAGAGATAG-TCT------AG------------------------------------------------TTCA--TGTATCCAATTGAGTCTAATAATTAAGTTATTCAACAGTAATGAT-----------AAACAGAAAA----------------------TCAAATCCCAAATTCGTTGACTA-GAG-TCC--------------TCA--------T---CTCCGGT--------------------------------------------------------------------------------------------GTCAAATCGACCCGCCCTCTTATACCCCGCCTCG-GT-CCAGAATCCTTCATCTAACTGCTCTAT-CC-T-CTCCCCC----------------------------------TATAGATATGTACCTACCCCTACCCAAAGACCCTTGTAGACCCTCGCCTTGTGGC--------------------- | |
| droTak1 | scf7180000415264:636184-636410 - | AAAAAAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AC------T-----ATTTAGAAATT-----------------------TATCTACTT---AG--AATTT---------------------------------------------TGAAGA----------------------AATTCCA-------------ATTTGA--AATAT-AAG-TTT--------------CCA--------T---TCCTGTT-----GCCCCAAT-----------------TTGCT-----TGG-------------------------------------------CATCCGAATAAATCGACCCACCCTTTCATACACTCCTC-CGT--CCAGCGTTCTTGATCTAACTGCTCTGC-CC-T-TCCTCCC----------------------------------CATAGATATGTACCTACCCCTGCCCAAAGACCCTTGTAGACCCTCGCCTTGTGGC--------------------- | |
| droEle1 | scf7180000491046:2172015-2172292 - | ATCCGGATTCACTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAATCCCTCTCAATACTGCCGGGAGATACCTAGATGTAGGATATTATTTCAC-------AT-AGGATGGGTCGAGGCTTACGGAAACGATGT------CATTAGTTATCAAAGAT-----------------------AGAAT---CGTTGA--AGTGATCCA-----------------------------------------------------------------------------------------------------------GAGAATTTC--------TTG---CGCTTTCAGTCG------------------------------------------------------------------------------------------AGTCCCTAAAAATCGACCCACTCTATAGTAATCCCAAT-TTT--CCGG----------ATAGCTCTTA---T--AA--CGATCACCCTGA-------------------------------CTTTTAG----TGACCGCACCTGTTGAACCGTGTCGTCCTTCGCCATGCGGT--------------------- | |
| droRho1 | scf7180000777227:322650-322932 - | ACCGGACTTCACCG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAATCCACTGCGCGCTTGCGTCGAAAGACCCTCAAGTATGACCCTATCCT------------GGGGTGGGTCGAATTGTATAGGAGAATAGT------AGC-------TAGAGTT-----------------------GGA---CTGAATTA--ATTTATTTA----------------------------AGGATGAA---------------------------------------------------------------------------ACATTCTTAGTCTACG------------------------------------------------------------TT-------------------------------------------CTTACAAACAAATCGGCCCACCCTTTTATACACCCTTT-AGA--CAAGAAGTATTGATCTAACTGCTATGA-CC-T--TTCCCA----------------------------------CATAGATATGTACCTGCCCCTGCCCAAAGACCCTTGTAGACCCTCGCCTTGTGGC--------------------- | |
| droFic1 | scf7180000454155:9320-9598 + | GCCTGGTTTCACCG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAATCCACTGCGCGCATGCGTCGAGAGGCCTTCAAGTATGACTCTGTCCT------------TGGGTGGGTCGACTTGTATAAAGAAAGGAT------TGCTA--------TGTT-----------------------GAG---CTTAATAG--AACTTTCAA----------------------------ATGACGAT-----------AAATAAGAAA-------------------------AAGC-------------------------------CTATTCCCC--------------------------------------------------------------T-------------------------------------------TCTCTAAGCAAATCGATCCACCCTTTTCT-TACCTCTC-AGA--C--------CCGACCTAATTGCCTTGT-CC-C--ACCCCA----------------------------------CATAGATATGTACCTGCCCCTGCCGAAAGACCCTTGTAGACCCTCGCCTTGTGGC--------------------- | |
| droKik1 | scf7180000302472:3199046-3199330 - | GCCAGGCTTCACTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAATCCCTTGCGGGCCTGTGTCGAGAGACCATCAAGTATGACTCTTTAGACC--------AAGGGAGGGATTGATCTGTAGGGAACTA-TGT------CC------------------------------------------------TTTC--TATATCCA-T----------------------------------------TGAGTA----------------------AATCCAT-------------CTTT---------CGTGTCTTGAGTCC--------TTGAAG---T--CT------TTAAAA--------------------------A----------TT-------------------------------------------CTTCCCCGCATATCAATCCCCCCCTC-------------T-TTCTATGGAGTTCCCATTTAATTGCTCCTT-CC-TTTCCCCTT----------------------------------CCTAGATATGTATCTGCCCCTGCCCCATGATCCATGTAGACCCTCGCCTTGTGGC--------------------- | |
| droAna3 | scaffold_12943:3314291-3314567 + | TCCCGGCTTCACCG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAATCCGCTGCGAGCCTGTGTGGAGAGACCTTCAAGTAGGAATTTAACCTCC-------CA-AGGGTGGTTCGATTTT--GGGTTAAAGAACCGCATATTC------GAAAAC--------------------------------TT---TA--AG-------------------------------------------------------------------------------TGGC-------------ATGT---------ATTGGCCAAACTCT--------------------CAGAT-----------------------------------TCTCT-----TTG-------------------------------------------CGAAATATCAAATCGACCCACCCTTTTGAAATCCCTA-----------AATTCCGTATCTAAACCCTCT--A--CTTCCCCACC----------------------------------CATAGACATGTACCTGCCCCTGCCCAAAGACCCTTGCAGACCCTCGCCTTGTGGG--------------------- | |
| droBip1 | scf7180000396433:209564-209844 + | GCCCGGTTTCACCG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAATCCGCTGCGAGCCTGTGTGGAGAGACCATCAAGTAGGAATCTGACCTCCA------CA-AGGGTGGTTCGATTTGG--GGTTAAAGAATCAAACATCCAAATT---------------------------------------------------------------------------------------------------TCCAG----------------------AAACGGC-------------ATTC---------ATTGGCCACATTCT--------CTAATA---T--T----------------------------------------TTCT-----TGG-------------------------------------------CAAACCCTTAAATCGACCCACCCTTCTGGAATCCCCAA-ACT--T--------CAGATCTAAACCCTGT--A--CTTCCCCACC----------------------------------CATAGATATGTACCTGCCCCTGCCCAAAGACCCTTGTAGACCCTCGCCTTGTGGG--------------------- | |
| dp5 | 4_group3:11342807-11343149 + | CCCCGAGTTTGAGGGAGCCCCGCCCAACTGCCGTCCCCAGTGTACATCCAGCTCTGAGTGTCCCTCCACCCAGGCTTGCATCAGCTACAAGTGTGCCGATCCCTGTCCTGGTGTGTGTGGCCAGCTTGCGATCTGTGAGGTGCGAAACCATAGTCCCATCTGTAGGTGTCCCCCGGCCATGATGGGCGATCCCTTCGTGCGCTGTC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGCCACGACCAGAGA----------------------GTAAGGAACGATCCAGAACTATGATCCAGAATGGTGTTAATCCTTTACGACCCTCG----------------------------------------------ACAGTT-----CC-T--CCACCC----------------------------------CTAAGAGATGTGGC--CCCCTAC--CGCGATCCCTGTGCACCCTCACCCTGTGGC--------------------- | |
| droPer2 | scaffold_8:2553761-2553964 + | ACCCAACTGTGAAT---------------------------------------------------------------------------------------------------------------------------------------------------------GCCTGCCCGGCTACACGGGAAACCCCATTGTCGGCTGTC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACCTGGTACCCGAGACGCCACGCTG-----TAAGA-------------------------------------------CCCCCACAAAGCTAGGCCC--CCGCT--GAAACCCTAC-TAA--TTGTCGCCATTTACCGCCCTC---------CT--GCTCGA----------------------------------TCTAGATCCCGACCCAATAGTACCCGAAAATCCATGCCAACCCTCGCCCTGCGGC--------------------- | |
| droWil2 | scf2_1100000004516:2703061-2703247 + | TACCGGTTTTACGG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGATGCCTTTACACGTTGCT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATCCC-----------------TTGCCAAGTAAGATAATTAA-----------------------------------------AATGGAATCCGACCAATTCGCATTACACTTT------------------------------------------------CTCCAATTTACAGTTCTCCCACCT----------ATTCAA----CAATTAGACGTTTCTCAGAACCCCTGCTTCCCGTCACCCTGCGGCCAGTATGCACAGTGCCGTGAT | |
| droVir3 | scaffold_12963:11804268-11804490 + | AGCGGGCATGACGG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGATCCGTTCCGACTCTGCCAGCAAGTGCCCCAAAGTAAGAACCAATCGGCCAACCCAACAAGTTTGCATATTGTATATAGGCAG-------G--------------------------------------------------------------------------G-----------------------------------------------------------------------C---------A--------------------------------------------------------------AC------------------------CTTTCT-----CCC----------CCAACCACCACAA--------------------------------------CAACTCTATATAA----------------------------------------------TCTTAAT-CTTACATA-TTCATTT------------ACAGCTTTGCCGAAACCATTGCCTACACCAAAGAATCCCTGCAATCCATCACCGTGTGGC--------------------- | |
| droMoj3 | scaffold_6500:20506719-20506952 + | TGCCGGTATGACGG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTGATCCGTTCCGACTTTGCCAGCAAGTTCAACAACGTAAGAACCAATTGTCCAACCAAACAAGTTTGCATATTGTATATAGGCAG-------G--------------------------------------------------------------------------G-----------------------------------------------------------------------C---------G--------------------------------------------------------------AC------------------------CTCTCT-----TC-------------------------------------------------------ACCACCCCACCCCACC------------CAA--CCCTAAATCTAGACCTAATTTAT----A--CA----------------------TTTTATATCGATTTCACAGCTGTGCCAACGCCAGTGCCTTCGCCGAAGAATCCATGCCAGCCATCGCCGTGTGGT--------------------- | |
| droGri2 | scaffold_15126:5321502-5321668 + | CCCAGGTTTCACAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAATCCCCTGCGTGCCTGCATCGAAAAGCCCACAAGTATACACACTCAAA------TAACAGATGTAGAATTTGTCTTTAAACAA--------------------------------------------------------------------------------------------------------------------------------------------------------TG-------------ACTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------CGTG---------------------------------------------T---ACCTGCC----------------------------------CTTAGATATGTACCTGCCATTACCCAAAGATCCATGCAGACCTTCACCCTGCGGC--------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 01:05 PM