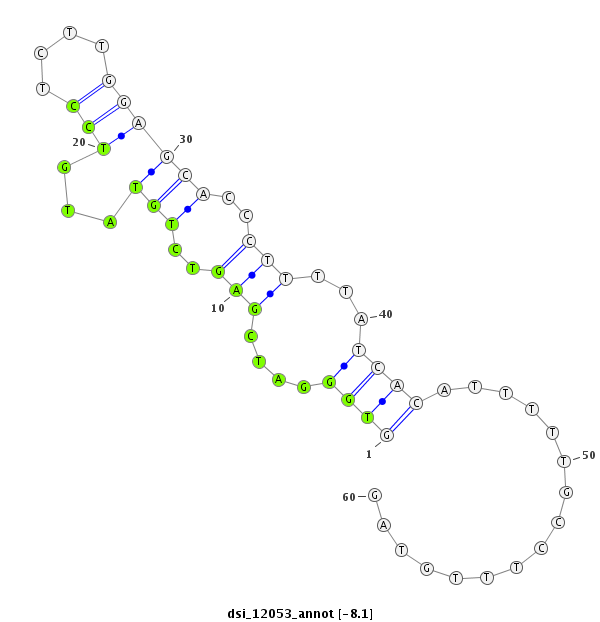
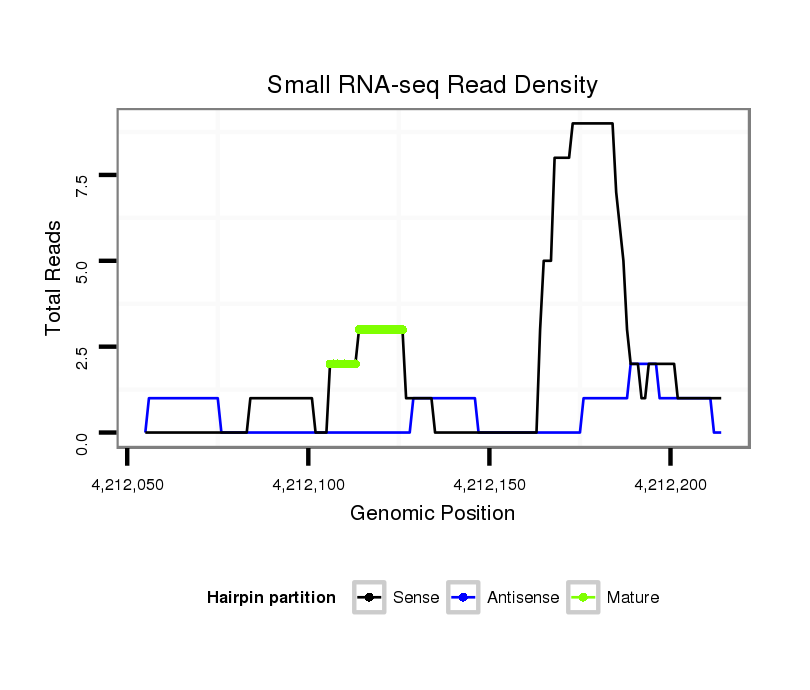
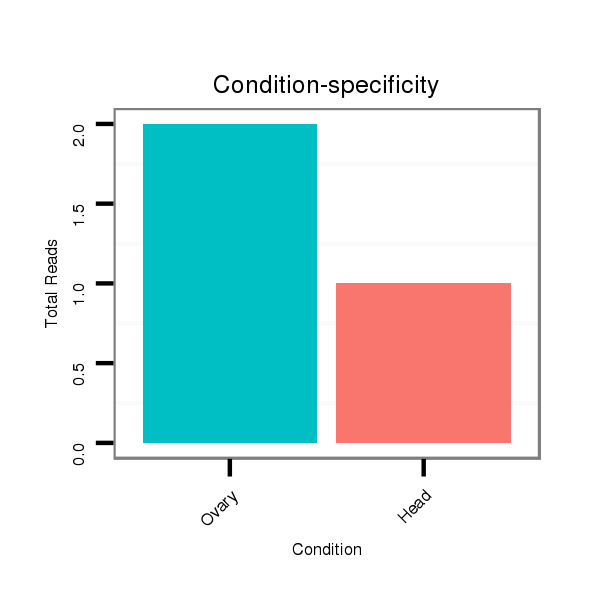
ID:dsi_12053 |
Coordinate:3r:4212105-4212164 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -7.2 | -7.1 |
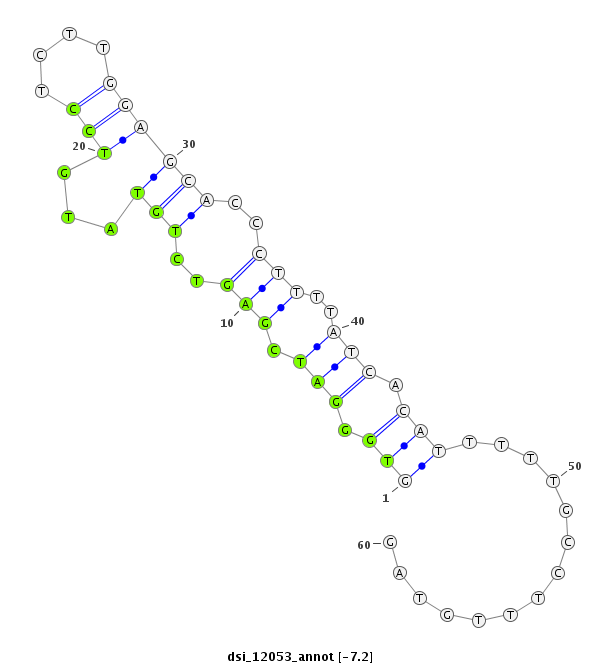 |
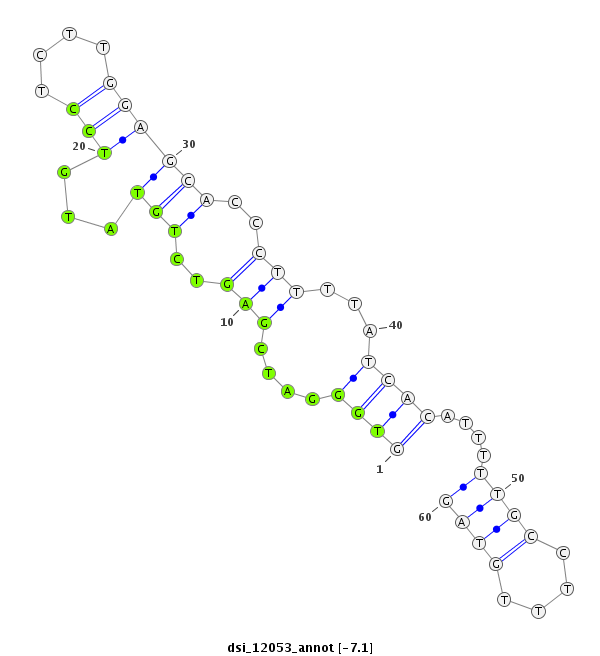 |
exon [3r_4211955_4212104_+]; CDS [3r_4211955_4212104_+]; CDS [3r_4212165_4212440_+]; exon [3r_4212165_4212640_+]; intron [3r_4212105_4212164_+]
No Repeatable elements found
| ##################################################------------------------------------------------------------################################################## TGCATGCCGTGGTCACAGTGCGTGATGCGAAGCACACCAACTTTGTGGAGGTGGGATCGAGTCTGTATGTCCTCTTGGAGCACCCTTTTATCACATTTTTGCCTTTGTAGTTCCGCAACTTCAAGATCGTCTACCGACGCTATGCGGGTCTGTATTTCTG **************************************************((((....(((..(((...(((....))))))..)))...))))................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
M025 embryo |
SRR553487 NRT_0-2 hours eggs |
M024 male body |
O002 Head |
SRR553488 RT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
M023 head |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................CCGCAACTTCAGGATCGTCTA........................... | 21 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................GTTCCGCAACTTCAAGATCGT.............................. | 21 | 0 | 1 | 2.00 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................CGCAACTTCAAGATCGTCTA........................... | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TGGGATCGAGTCTGTATGTCC........................................................................................ | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................CTATGCGGGTCTGTATTTCTG | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................AGTCTGTATGTCCTCTTGGAG................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CTTCAAGATCGTCTACCGACGCTATGCGG............. | 29 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................CGCAACTTCAAGATCGTCTAC.......................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................GTTCCGCAACTTCAAGATCGTC............................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............ACAGTGCGTGATGCCAAGCACAC........................................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................TTCCGCAACTTCAAGATCGTCTACCGA....................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................AAGCACACCAACTTTGTG................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TTCCGCAACTTCAAGATCGTCT............................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................CCGCAACTCCAAGATCGCCT............................ | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................TTCTAGTTCGTCTACCGAC...................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................GCCTTTGTAGTTCCGTT........................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................CTTAGTGTAGGTGGGA........................................................................................................ | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
|
ACGTACGGCACCAGTGTCACGCACTACGCTTCGTGTGGTTGAAACACCTCCACCCTAGCTCAGACATACAGGAGAACCTCGTGGGAAAATAGTGTAAAAACGGAAACATCAAGGCGTTGAAGTTCTAGCAGATGGCTGCGATACGCCCAGACATAAAGAC
**************************************************((((....(((..(((...(((....))))))..)))...))))................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
O001 Testis |
O002 Head |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|
| .CGTACGGCACCAGTGTCACG........................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..TTACGGCACCAGTGTCACGCAC........................................................................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................AACCTCGTGGGAAAATAG.................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................GTTCTAGCAGATGGCTGCGAT.................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................GCTGCGATACGCCCAGACATAAA... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .TGTACGGCACCAGTGTCACG........................................................................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................TCGCGGGAACATAGTATAAAAA............................................................ | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................TGAGATATGCAGGAGAAC................................................................................... | 18 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| .......................CAAAGCTTCGTGTGGTT........................................................................................................................ | 17 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................GATATGCAGGAGAACTTC................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/23/2015 at 11:19 PM