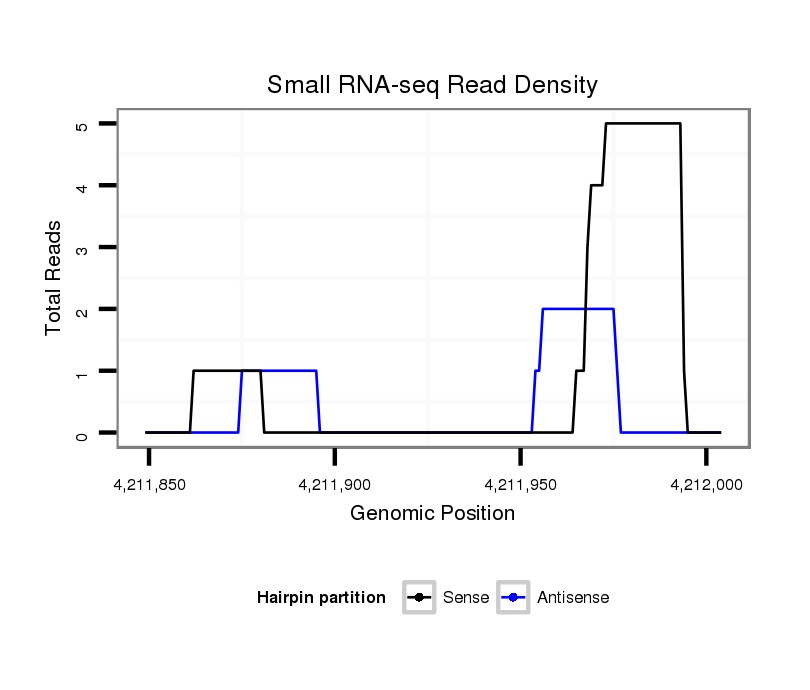
ID:dsi_11934 |
Coordinate:3r:4211899-4211954 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
CDS [3r_4211896_4211898_+]; five_prime_UTR [3r_4211772_4211895_+]; exon [3r_4211772_4211898_+]; exon [3r_4211955_4212104_+]; CDS [3r_4211955_4212104_+]; intron [3r_4211899_4211954_+]
No Repeatable elements found
| ##################################################--------------------------------------------------------################################################## AAAAAGTTGTAAATAAGCAGGAAACGGTCTAACCAATTTCCCGAAAAATGGTAAGCAACTAGCCAGCTATAAGCTTAGACCCCTATTTGACCCTCCATTTGCCCAGATACGATTCATACTTATACAAAACCGAGCTGGAAAAACTCGCCTGGCCAA **************************************************.(((((...(((....)))...))))).............................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
M023 head |
M024 male body |
M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
GSM343915 embryo |
SRR618934 dsim w501 ovaries |
SRR902008 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................TTATACAAAACCGAGCTGGAAAAACT........... | 26 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................TATACAAAACCGAGCTGGAAAAACTC.......... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................TACTTATACAAAACCGAGCTGGAAAAACT........... | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................CAAAACCGAGCTGGAAAAACT........... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................AAACCGAGCTGGAAGGACTC.......... | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............TAAGCAGGAAACGGTCTAA............................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................TGGGAAGCAACTAGC............................................................................................. | 15 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........GTGCATAAGTAGGAAACGG................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................CCCGAAAGATGGTAAC..................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TTTTTCAACATTTATTCGTCCTTTGCCAGATTGGTTAAAGGGCTTTTTACCATTCGTTGATCGGTCGATATTCGAATCTGGGGATAAACTGGGAGGTAAACGGGTCTATGCTAAGTATGAATATGTTTTGGCTCGACCTTTTTGAGCGGACCGGTT
**************************************************.(((((...(((....)))...))))).............................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
M023 head |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M024 male body |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................CTATGCTAAGTATGAATATGTT............................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................CCAGAATTGTTAAAGGGCT................................................................................................................ | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................CAGATTGGTTAAAGGGCTTTT............................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................ATGCTAAGTATGAATATGTTT............................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................ATGGGTCGATGTTCGAA................................................................................ | 17 | 2 | 20 | 0.65 | 13 | 0 | 0 | 0 | 13 | 0 | 0 | 0 |
| .........................................................................................................................TATGTTTTCGCTAGACCT................. | 18 | 2 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ..........................CAGAATTGTTAAAGGGCT................................................................................................................ | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................CCATTGCCAGAATTGTTAA...................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| .................................................................................................................AGTATGAAAATAATTTGGCT....................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/23/2015 at 11:13 PM