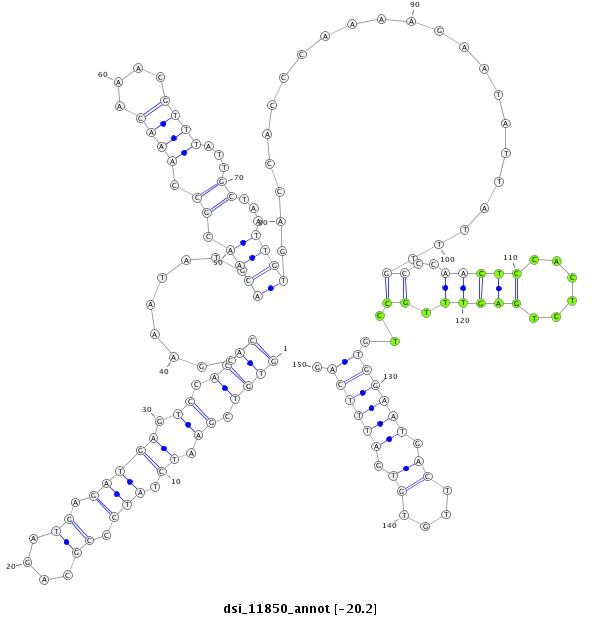
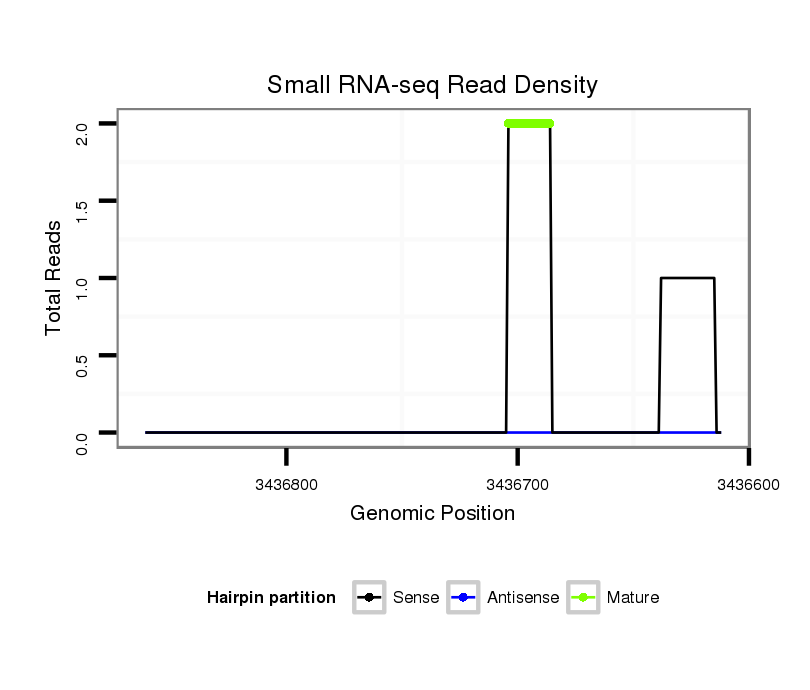
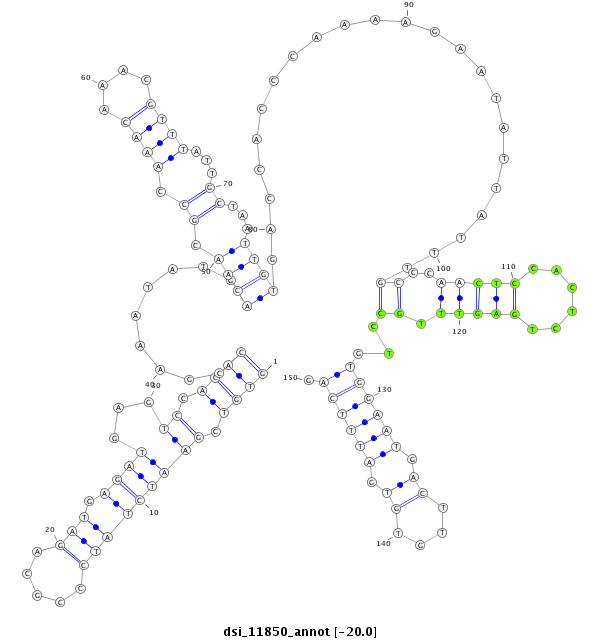
ID:dsi_11850 |
Coordinate:2r:3436662-3436811 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
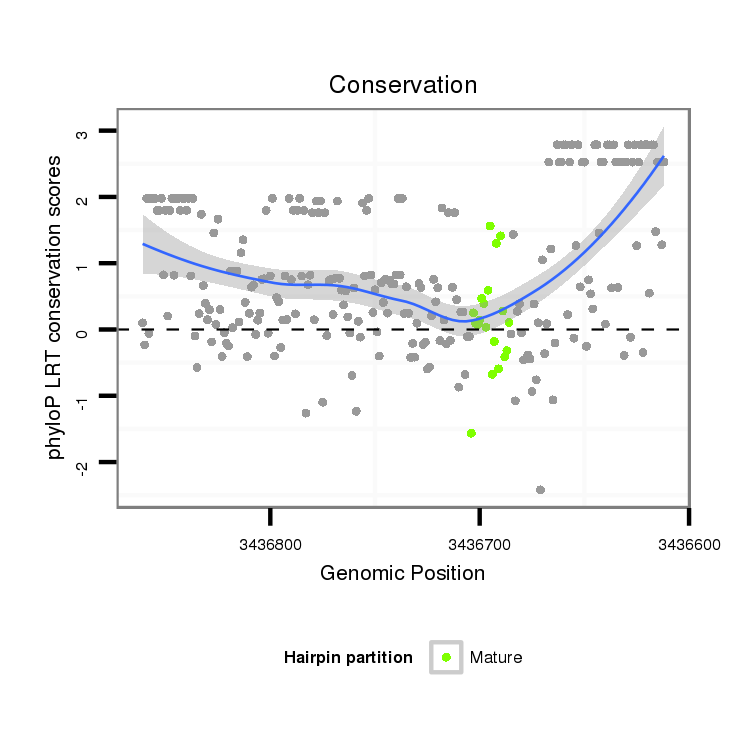
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -20.0 | -19.7 | -19.4 |
 |
 |
 |
CDS [2r_3436535_3436661_-]; exon [2r_3436535_3436661_-]; intron [2r_3436662_3437013_-]; Antisense to intron [2r_3425861_3441727_+]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGTATTTGCTTGCGATTAGTTGTTAAAGCAAGGGTTTCGGTGCTCCGTGTGTGTCGAATCTATCCCGCAGATGAGATGAGTCCACACCGAAATATGACAACGCCAAACAAACGTTTATTGCTAATTGTGACCACCCAAAAGAATATTATTTGCCCAACTCCACTCTGAGTTTGCCTGTGGAATGACTTGTGTGATTTCAGGTTCGTGCTGCGATCCTATGGGATTTCCGGGCTGCAGCATTACATACGTC **************************************************((((.((.((.(((.((....)).))))).)).)))).........((((.((.((((....))))...))...)))).......................((..(((((......))))).))...((((((.((....)).)))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR618934 dsim w501 ovaries |
GSM343915 embryo |
M025 embryo |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR902009 testis |
SRR553485 Chicharo_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................CTCCACTCTGAGTTTGCCT.......................................................................... | 19 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................TTTCCGGGCTGCAGCATTACATAC... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................GATTGCAGGTTCGTCCTG........................................ | 18 | 2 | 9 | 0.67 | 6 | 2 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| TGTATTTGCTTGCGA........................................................................................................................................................................................................................................... | 15 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................AGGTTAGTGCTACGATCCCAT............................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................GTGTGGATCGAATCTATC.......................................................................................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................GATTGCAGGTTCGTG........................................... | 15 | 1 | 17 | 0.29 | 5 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................GTTGTTAGAGCAAGG......................................................................................................................................................................................................................... | 15 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................TGTGTCGAATCTGCCCCG....................................................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................AAAGATTTTTATTGGCCCAAC............................................................................................ | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................ACTCTGAGATTGCCTGT........................................................................ | 17 | 1 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................CTATCCAGCAGATGATA.............................................................................................................................................................................. | 17 | 2 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................ACACGGTAATAGGACAACG.................................................................................................................................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........TGCGATTAGTTCTAAAAG.............................................................................................................................................................................................................................. | 18 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................GATTGCAGGTTCGTCCGG........................................ | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
ACATAAACGAACGCTAATCAACAATTTCGTTCCCAAAGCCACGAGGCACACACAGCTTAGATAGGGCGTCTACTCTACTCAGGTGTGGCTTTATACTGTTGCGGTTTGTTTGCAAATAACGATTAACACTGGTGGGTTTTCTTATAATAAACGGGTTGAGGTGAGACTCAAACGGACACCTTACTGAACACACTAAAGTCCAAGCACGACGCTAGGATACCCTAAAGGCCCGACGTCGTAATGTATGCAG
**************************************************((((.((.((.(((.((....)).))))).)).)))).........((((.((.((((....))))...))...)))).......................((..(((((......))))).))...((((((.((....)).)))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
M023 head |
|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................TGAGGTTAGATTGAAACGGA.......................................................................... | 20 | 3 | 11 | 0.27 | 3 | 2 | 0 | 1 | 0 |
| ...............................................ACACACAGCTTACATA........................................................................................................................................................................................... | 16 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 |
| ..................................AAAGCCACGAGTCACA........................................................................................................................................................................................................ | 16 | 1 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................TCAGAAAAGTCCAAGCACG.......................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 |
| .................................................................................................GGTGCGGTTTGTTTGAAGAT..................................................................................................................................... | 20 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ........................................ACACGGCACACACAGCT................................................................................................................................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ........................................................................................................................GATTAATACTGGTTGGT................................................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 2r:3436612-3436861 - | dsi_11850 | TG---TATTTGCTTGCGAT-TAGTTGTTAAAGC-------------------------AAGGGTT-TCGGTGCTCCGTGTGTGTC---GAATCTATCCC------GCAGATGAGATGAGTCCACACCGAAA---TATGA-CAA------CGCCAAACA--------AACGTTT-----ATTGCTAATTGTG------------------------ACCACCCAAAA-----------------------------------G----------AATATTATTTG--CCC-AA-CTCCACTCTGAGTTTGCC--TGTGGAATGACTTGTGTGATTTCAGGTTCGTGCTGCGATCCTATGGGATTTCCGGGCTGCAGCATTACATACGTC |
| droSec2 | scaffold_1:238422-238671 - | TG---TATTTGCTCGCGAT-TAGTTGTTAAAGC-------------------------AAGGGTT-TCGGTGCTCCGTGTGTGTC---GAATCTATCCC------GCAGATGAGATGGGTCCACACCGAAA---TATGA-CAT------CGCCAAACA--------AACGTTT-----ACTAGCAGTTGTG------------------------ACCACCCAAAA-----------------------------------G----------AATATTATTTG--CCC-AA-CTCCACTCTGAGTTTGCC--TGTGGAATGATTTGTATGATTTCAGGTTCGTGCTGCGATCCTATGGGATTTCCGGGCTGCAGCATTACATACGTC | |
| dm3 | chr2R:2574242-2574491 - | TG---TATTTGCTTGCGAT-TAGTTGTTAAAGC-------------------------AAGGGTT-TCGGTGCTCCGTGTGTGCC---GAATCTATCCC------GCAGATGAGATGAGTCCACATCGAAA---TATGA-CAA------CGCCAAACA--------TACGTTT-----ATTGCTAATTGTG------------------------ACCACCCAAAA-----------------------------------G----------AATATTATTTG--CCC-AA-TTCCACTCTGAGTTTGCC--TGTAGAAAGACTTGTATGATTTCAGGTTCGTGCTGCGATCCTATGGGATTTCCGGGCTGCAGCATTATATACGTC | |
| droEre2 | scaffold_4929:19971521-19971776 + | TG---TATTTGCTTGCGATTTAGTTGTAAAAAC-------------------------AAGGGTT-TCGATACCCCGTGTGTGCC---GAATCTATCCC------GCAGATGAGATGGGTCCACACCAAAA---TATGA-GAA------CGCCAAACA--------AACGATT-----ATTGCTAATCGTG------------------------ACCACCCAATGGA--------------------------CCA---GG----------AATATTATTTA--CCC-CT-ATCCATTCTGTGTGTGCC--TCT-GAATGACTTGTATGATTTCAGGTTCGTGCTGCGATCTTATGGGATTTCCGGGCTGCAGCACTACATACGTC | |
| droYak3 | 2L:15317464-15317695 - | TG---TATTTGCTTGCGACTTAGTTGTAAAAGC-------------------------AAGGGTT-TCGATACCCCGTGTATGCC---GAATCTATACC------GCAGATGAGATGAGA----------------------A------CGCCAAACA--------AACGTTT-----ATTGCTAATTGTG------------------------AGCACGGCAAA-----------------------------------A----------AATACTATTTA--CCC-AA-ATCCATTCTGTATGTGCC--TCT-GAATGGCGTATATGATTTCAGGTTCGTGCTGCGATCTTATGGGATTTCCGGACTGCAGCATTACATACGTC | |
| droEug1 | scf7180000409462:3446183-3446455 - | CG--GTATTTGCTTGCGATTTAGTTGTAAATAC-------------------------CAGGTTT-TCTAAACCCCATGAACGCT---GGATCTATCCC------TCAGATGAGAAGAGTCCACAACGAAA---TATGA-CATG-ACA-TGACGAGCA--------AACAT---GGTCATTGCTAATTGTT------------------------CTCACCCAAAAGAATCTAGCAAGCTTAAGCACTTAAGAACGC---TG----------AATACTATTTC--CCA-AA-AT-------------G-----CT-GAACGAAGAACGTATTTATAGGTTCGTGCTGCGATCTTATGGGATTTCCGGGCTGCAGCACTACATACGTC | |
| droBia1 | scf7180000302292:3577792-3578031 + | CG---TATTTGCTTGCGATTTAGTTGTAAACAC-------------------------A------------------TGTGTGCC---GAATCTATCCC------GTGGATGAGATGTGTCCACATCGAAA---TATGA-CAT------GGTCAAACATCAACATCAACTCCT-----ATTGCTAATTGTT------------------------CCCATCCCAAAGACA-------------------------CC---CG----------AATAATGCTTA--------------TTGTGAGTGTGCATGCCT-GACCAAAGATCGCCTTTTCAGGTTCGTTCTGCGTTCTTATGGGATTTCCGGGCTTCAGCACTACATACGTC | |
| droTak1 | scf7180000413887:28881-29104 + | CT---TGTTTGCTTGTGATTTAGTTGTAAGCAC-------------------------------------------ATGTGTGCC---GAATCTATTCC------CCAGATGAGATGGGTCCACACCGAAA---TATGA-CAT------GGTCAAACATC------AACATTT-----ATTGCTAATTGTT------------------------CAC---------------------------------GAACCC---CG----------AATAATATTTA--------------TTCCGAGTGTGTA--CCT-CAACGAAAATCATATTTTCAGGTTTGTTCTGCGATCTTATGGGATTTCCGGACTCCAGCACTACATACGTC | |
| droEle1 | scf7180000491217:441044-441270 + | CG---TATTTGCTTGCGATTTAGTTGTAA----------------------------------------------------TGCC---GAATCCATTGC------CCAAATGAGATGAGTCCACACCGAAA---CACGA-CGAA-ACACGGGCGAACA--------AACATTT-----ATTGCTAATTGTTG-----------------------CCCACCCAGAAGACACC--------------------AACTA---CG----------AATTTTATTTA--TTT-A--------------TATG----TCT-GAAGTAAGATCGTATTTACAGGTTCGTGCTGCGATCTTATGGGATTTCCGGGCTGCAGCATTACATACGTC | |
| droRho1 | scf7180000766409:461392-461674 - | CG---TATTTGCTTGCGATTTAGTTGTAAGTAC-------------------------AAGGGTC-TCTATGCCCCATGTATGCT---GAATCCATCGC------CCAGATGAGATGAGTCCACACCGAAA---TATGA-CAT------GGTCGAACA--------AACATTT-----ATTGCTAATTCTTT-----------------------CCCACCCAGAAGACAATAACAA-C--TAGTACTTAAGAACGA---CG---------AAATATTATTTC--CCA-AA-TTCGTTTCTGTGTGTGTATGTTT-GTACGTAGATCATATTTATAGGTTCGTGCTGCGATCTTATGGGATTTCCGGACTGCAGCACTACATACGTC | |
| droFic1 | scf7180000453955:727900-728160 + | CG---TGTTTGCTTGTGATTTAGTTGTAAGCAC-------------------------AGGGGTT-TTCATGCCCCATGTGTGCC---AAATCTGTCCC-----CCCAGATGGGATGAGTCCACACCGAAA---TATGA-CAT------GGGTAAACA--------AGCGTTT-----ATTGCTAATTGCT------------------------CCCACCCAGAAGAAGA----AA-C--T----------AATCC---CG----------AGTGTTGTTCA--CCA-AAAAAACATTCTGTAT--------CT-GAACGTCGATCGTATTTTTAGGTTCGTGCTGCGATCTTATGGGATTTCCGGGCTGCAGCACTACATTCGTC | |
| droKik1 | scf7180000302470:137135-137384 - | CA---TATTTGCTTGCGATTTAGTTGTAAAAAC-------------------------AAAACTTTTCCA-----------TGCT---CCAGCCATCCG------CGAAATGAGATGGGTCCACAGCAAAGACTTATGA-CAT------AAGCAAACA--------AGGAATT-GATAATTGCTAATTGGC-------AACTCTACTCCATAGACTTTACAAAACAGAGA---------------------------------CAGTTT------------------------------TTGCGTTTCTT--TCT-GACCAAAAAGTATCTTTCTAGGTTCGTGCTGCGTAGTTATGGAATCTCCGGGCTGCAGCACTACATACGTC | |
| droAna3 | scaffold_13266:18435599-18435814 + | -G---TGTTTGCTTGCGATTTAGTTGTAAAAAC-------------------------A----------------------CTCC---GAATCCATTCCGGAGCCCCAAACGCGATGGGTCTACATCGGAG---TATAT-CTT------CGGCAAACA--------AACATTC-----GCTGCTAATTGTGTT--------------ACA---AAACGGCTTAGAAG---------------------------------CGAATATCTTTGGGT----------------------TTCTGT----------------------TTCTGTTTGCAGGTTCGTGCTGCGGAGTTATGGGATTTCCGGACTACAGCACTACATACGTC | |
| droBip1 | scf7180000396735:1707635-1707855 + | -G---TGTTTGCTTGCGATTTAGTTGTAAAAAC-------------------------A-----T-TCCGGG---------CTCC---GAGTCCATTCCAGAGCCCCAAATGAGATGGGTCTACATCGGAG---TAT------------------ACA--------AACACTC-----GTTGCTAATTGTATT--------------ACA---AAACGACTCGCAA-------------------------GAACCAACGCGAATATTTTTGAATATTATT------------------------GTG-------------------GTCTTTGCAGGTTTGTGCTGCGATCTTATGGGATTTCCGGACTACAGCACTACATACGTC | |
| dp5 | 3:839558-839840 - | TTCATTATTTGCTTGCGATTTAGTTGTAAAAACATAGAAATCAAACTTTGGAAAGAGCAAAGGTC-T-----------GTGTGCCGCGGAATCTATC---------CAAATGAGATGAATCAACACCAAGA---TATGTGCATGTGTA-GGTAGAACA--------AACATTCA----ATGGCTAATGGTTTCCTGTCCGATC-ATTCCATC-AGTCCACTCGAAATATA---------------------------------CA-------ATTATTGTTTTTGCCGGCT-TTCCATGCTG------------------------TTTCCTTGCAGGTTTGTGCTGCGATCCTACGGGATTTCCGGACTACAGCACTACATACGTC | |
| droPer2 | scaffold_2:1015491-1015773 - | TTCATTATTTGCTTGCGATTTAGTTGTAAAAACATAGAAATCAAACTTTGGAAAGAGCAAAGGTC-T-----------GTGTGCCGCGGAATCTATC---------CAAATGAGATGAATCAACACCAAGA---TATGTGCATGTGTA-GGTAGAACA--------AACATTCA----ATGGCTAATGGTTTCCTGTCCGATC-ATTCCATC-AGTCCACTCAAAATATA---------------------------------CA-------ATTATTGTTTTTGCCGGCT-TTCCATGCTG------------------------TTTCCTTGCAGGTTTGTGCTGCGATCCTACGGGATTTCCGGACTACAGCACTACATACGTC | |
| droWil2 | scf2_1100000004954:1996515-1996570 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCTAGGTTTGTATTGCGTTCTTATGGCATATCTGGCCTCCAACATTATATACGAC | |
| droVir3 | scaffold_12875:15456237-15456289 - | ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGTTTGTGCTGCGCTCGTATGGAATATCCGGACTACAGCATTATATACGAC | |
| droMoj3 | scaffold_6496:8339963-8340023 + | TA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCATCTCAGGTTTGTCCTACGCTCTTATGGCATATCCGGACTACAGCACTATATACGAC | |
| droGri2 | scaffold_15245:12259344-12259396 - | ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGTTTGTTCTACGCTCTTATGGCATATCTGGACTACAGCATTATATACGAC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 10:49 AM