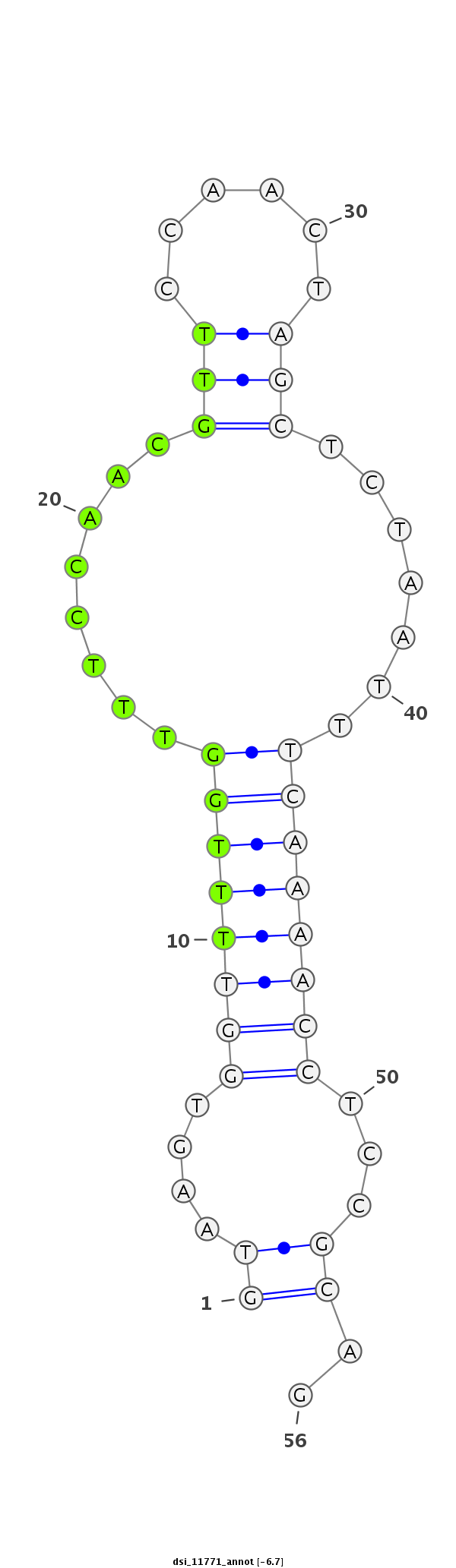
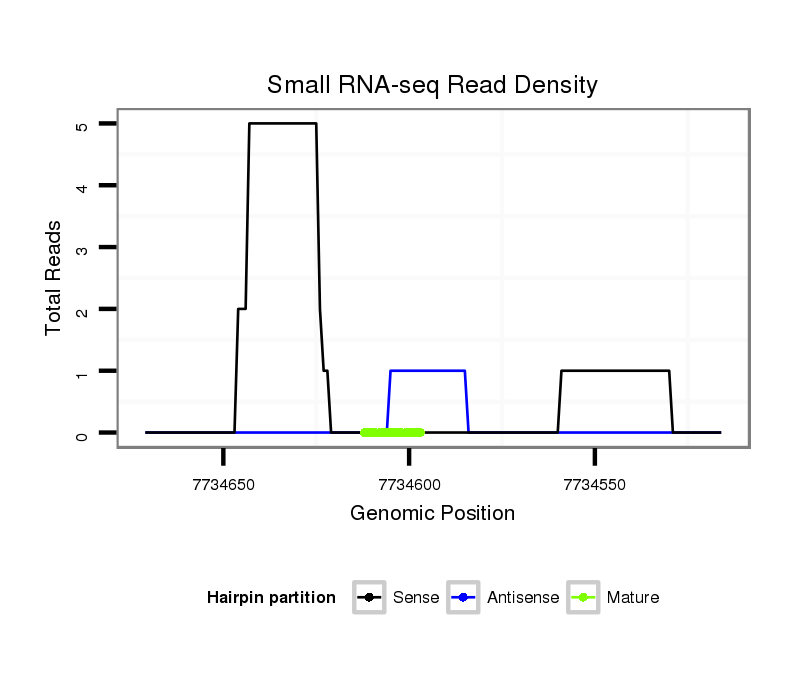
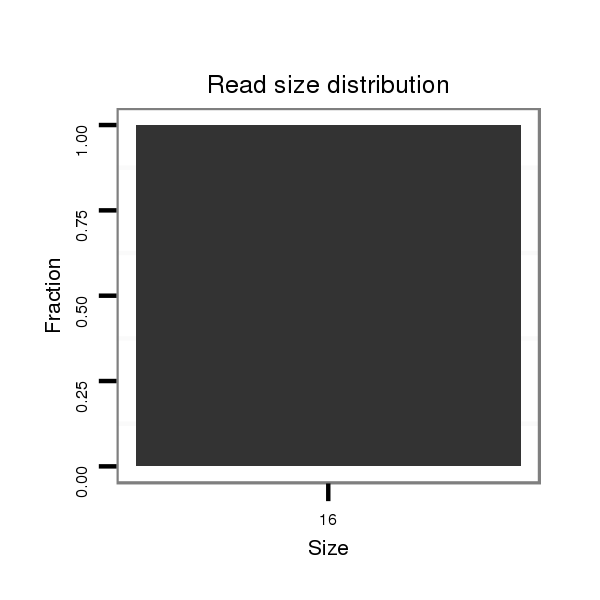

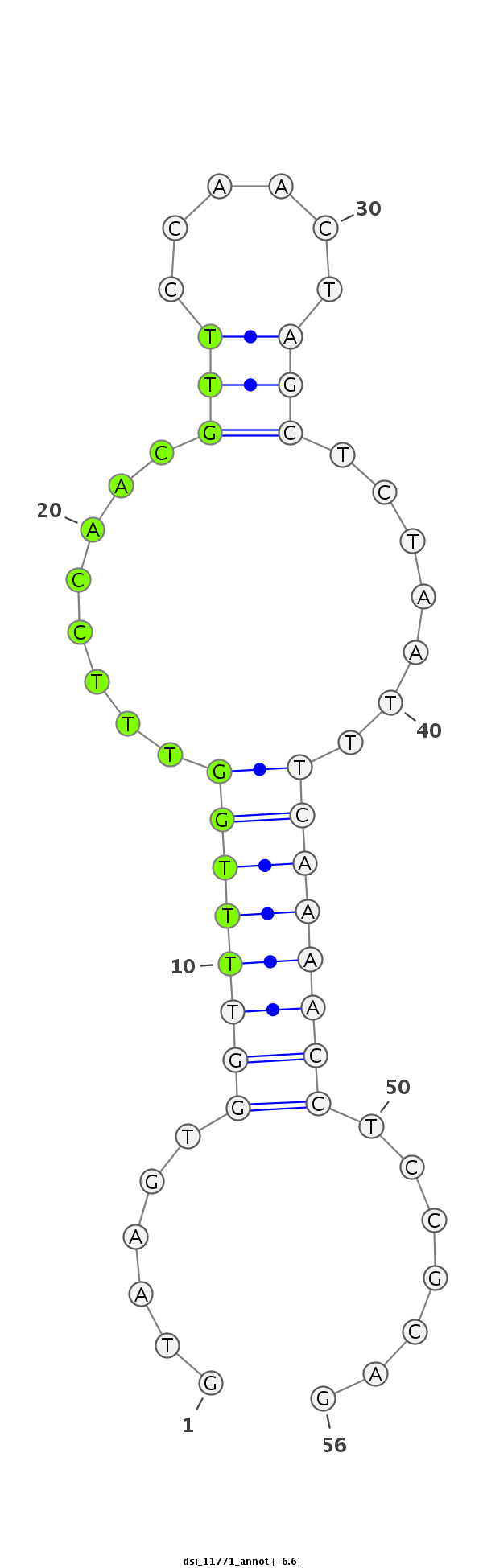
ID:dsi_11771 |
Coordinate:3l:7734566-7734621 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -6.6 | -6.2 | -6.1 |
 |
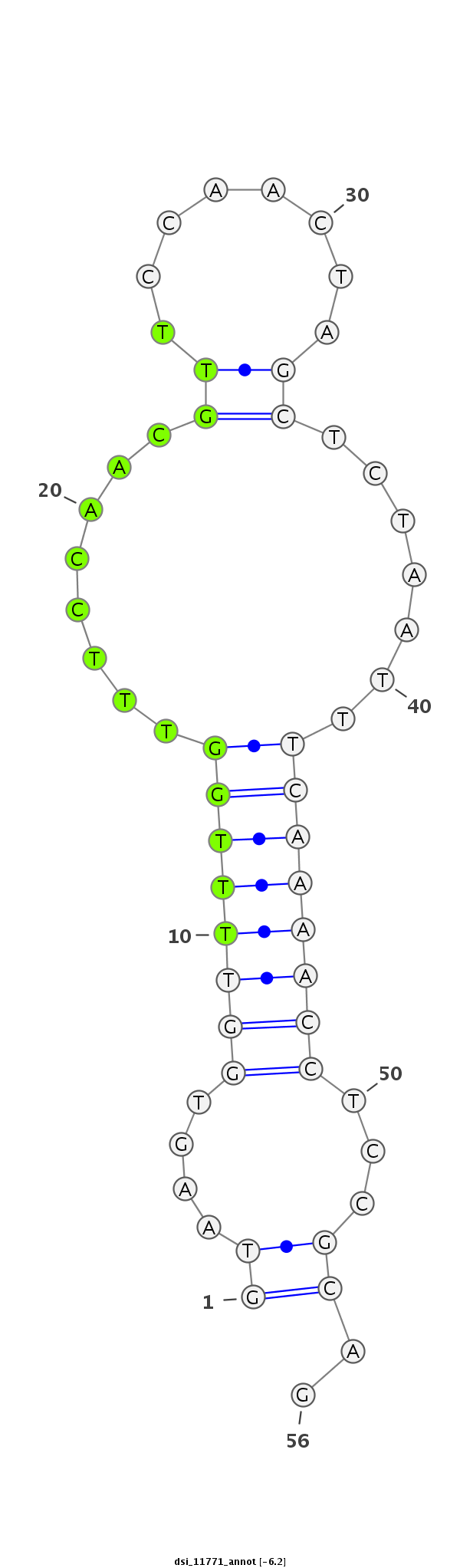 |
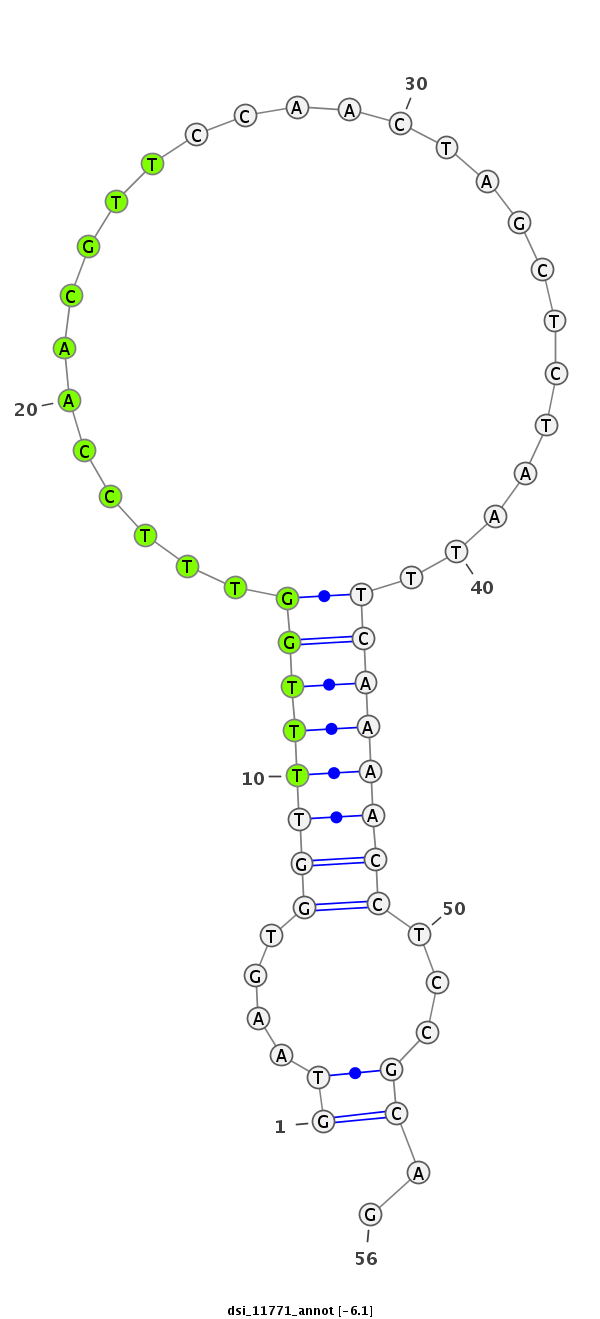 |
exon [3l_7733973_7734565_-]; CDS [3l_7733973_7734565_-]; CDS [3l_7734622_7734947_-]; exon [3l_7734622_7735032_-]; intron [3l_7734566_7734621_-]
No Repeatable elements found
| mature | star |
| ##################################################--------------------------------------------------------################################################## GTGAAGCTCCGCCGGCGCGTCCAAATGTACCAGTGGGTCGAGGAGACGGTGTAAGTGGTTTTGGTTTCCAACGTTCCAACTAGCTCTAATTTCAAAACCTCCGCAGCGAGCACAACTATGGCGACAGCGTGGGAACGACGCAGTCGGACTCCCGCA **************************************************((....((((((((........(((......))).......))))))))...))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
M023 head |
M025 embryo |
SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M053 female body |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................GTACTAATCGGTCGAGGA................................................................................................................ | 18 | 3 | 20 | 12.95 | 259 | 170 | 74 | 2 | 0 | 0 | 0 | 0 | 13 | 0 |
| ............................ACCAGTGGGTCGAGGAGAC............................................................................................................. | 19 | 0 | 1 | 3.00 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................TGTACCAGTGGGTCGAGGAGACGGT.......................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................TGTACCAGTGGGTCGAGGAGACG............................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................CAACTATGGCGACAGCGTGGGAACGACGCA.............. | 30 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................TTTGGTTTCCAACGTT................................................................................. | 16 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................GCGACAGCGTGGGGA..................... | 15 | 1 | 10 | 0.20 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................GCTTGGGAACACCGCAGTCG.......... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................GAACGACGGACTCTGACTC..... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CACTTCGAGGCGGCCGCGCAGGTTTACATGGTCACCCAGCTCCTCTGCCACATTCACCAAAACCAAAGGTTGCAAGGTTGATCGAGATTAAAGTTTTGGAGGCGTCGCTCGTGTTGATACCGCTGTCGCACCCTTGCTGCGTCAGCCTGAGGGCGT
**************************************************((....((((((((........(((......))).......))))))))...))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR902009 testis |
SRR553486 Makindu_3 day-old ovaries |
M025 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................GCGTCAGACTGAGGGC.. | 16 | 1 | 1 | 5.00 | 5 | 4 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................AGGTTGCAAGGTTGATCGAGA..................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................GCGTCAGACTGAGGGCCT | 18 | 2 | 5 | 0.40 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ................................................................AAAGGCTGTAAGGTTTATCGAG...................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................GGATTGAAGTTTTGGAGG...................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................CGTCAGACTGAGGGC.. | 15 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................GCGTCAGACTGAGGGCC. | 17 | 2 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................AGCGTCAGACTGAGGGC.. | 17 | 2 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................CCTGCCACATTCTCCAGAA.............................................................................................. | 19 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................CATAAGTTGTAAGGTTGATC......................................................................... | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................GCGTCAGACTGAGGGCT. | 17 | 2 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AAAAATTGCAAGGTTGAT.......................................................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................GGCGTCAGACTGAGGGCCT | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................AAGGTTGCGAGGTTG............................................................................ | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................CTCGTGATCATACCGC................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
Generated: 04/24/2015 at 10:31 AM