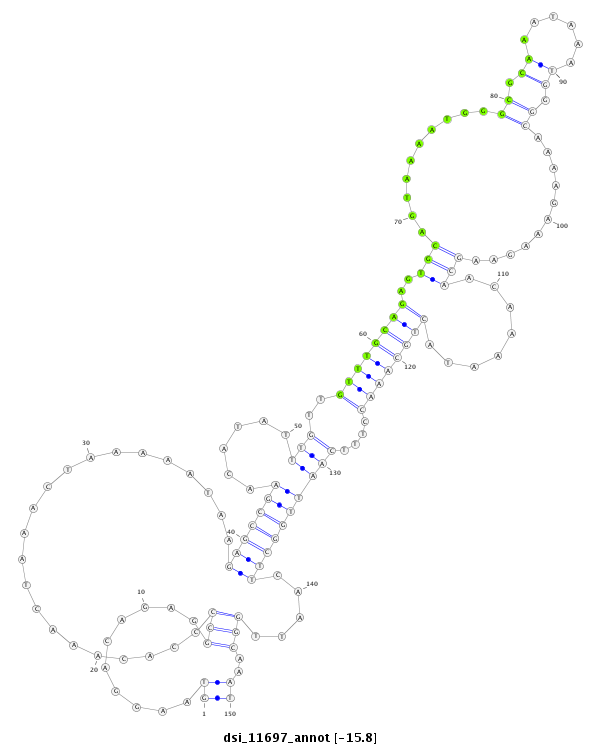
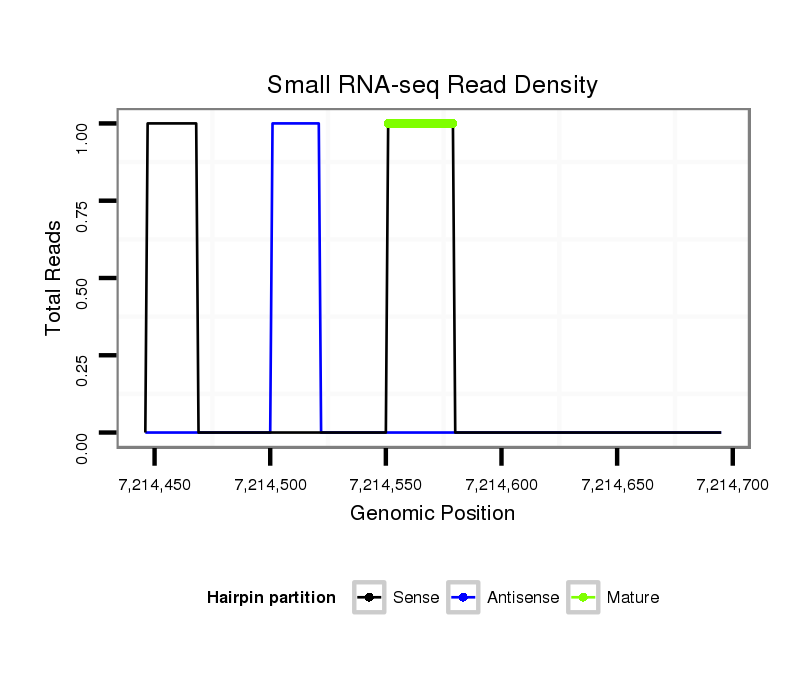
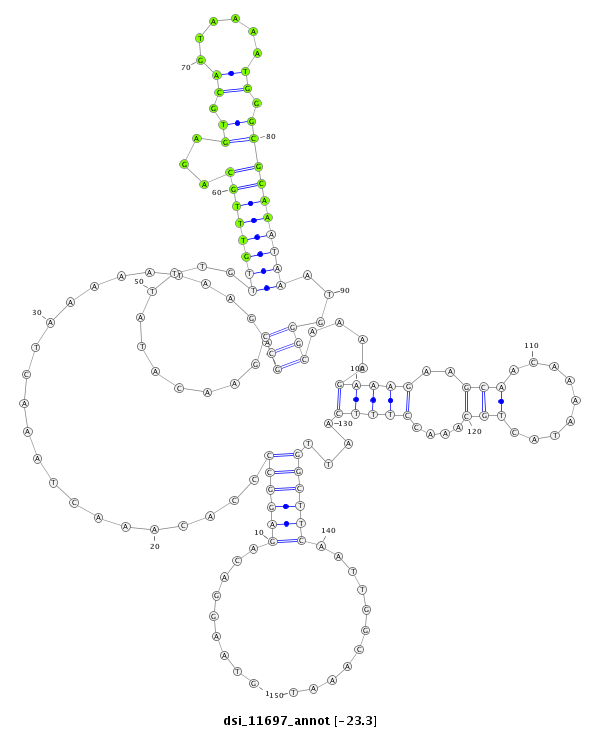
ID:dsi_11697 |
Coordinate:2r:7214496-7214645 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -23.3 | -23.3 | -23.2 | -23.2 |
 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CAGGCGGCAAATTGCAGCAGCAGCGAAAATCAAATATAAATAGCGAAAAGGTAAGGACAGAGGCCCCACAAACTAAACTAAAAATAAGAGCCGAACATATTTGTTGTTTGCAGAGTGCAGTAAAATGGGCGCAAATAAATGGGCAAAAGAAAGAAGCAACAAAATACTGCAAACCTTTCAATTGGCTTCAATTGGCAAATGCAATTGTTCAGCATTACATTTGCATTACACCGCTCTCCAACTCTTGCTC
**************************************************((..........(((......................(((((((......(((..((((((((..(((..........((.((......)).))...........)))........))))))))....)))))))))).....)))..))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
M053 female body |
GSM343915 embryo |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................GTTTGCAGAGTGCAGTAAAATGGGCGCAA.................................................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .AGGCGGCAAATTGCAGCAGCAG................................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................GGGCAAAAGAAACAAGCAT........................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 |
| .................................................GGTAAGGGAAGAGGCCC........................................................................................................................................................................................ | 17 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................TAAATGGGCATAAGAAATA................................................................................................ | 19 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CAATTGTTGAGCAGTACATG............................. | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 |
| ...............................................ATGGTAGGGACAGAGGGC......................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
GTCCGCCGTTTAACGTCGTCGTCGCTTTTAGTTTATATTTATCGCTTTTCCATTCCTGTCTCCGGGGTGTTTGATTTGATTTTTATTCTCGGCTTGTATAAACAACAAACGTCTCACGTCATTTTACCCGCGTTTATTTACCCGTTTTCTTTCTTCGTTGTTTTATGACGTTTGGAAAGTTAACCGAAGTTAACCGTTTACGTTAACAAGTCGTAATGTAAACGTAATGTGGCGAGAGGTTGAGAACGAG
**************************************************((..........(((......................(((((((......(((..((((((((..(((..........((.((......)).))...........)))........))))))))....)))))))))).....)))..))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
SRR553488 RT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M053 female body |
SRR902008 ovaries |
M023 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................GTCAGGTGGCGAGAGGTTGA....... | 20 | 2 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................ACAAACGTCTCACGACA................................................................................................................................. | 17 | 1 | 2 | 2.00 | 4 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................TGTCAGGTGGCGAGAGGTTGA....... | 21 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................GTCAGGTGTCGAGAGGTTGA....... | 20 | 3 | 11 | 1.82 | 20 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................AAGCACAAACGTCTCACGACA................................................................................................................................. | 21 | 3 | 3 | 1.67 | 5 | 0 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................CTGTCTCCGGGGTGTTTGATT.............................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......GTTTAACGTCGTCGCTGCATTTAGT.......................................................................................................................................................................................................................... | 25 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................AAAGCACAAACGTCTCACG.................................................................................................................................... | 19 | 2 | 5 | 0.80 | 4 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ..............................................................................................................GTCTCACGTCATGGTAGCCGC....................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................CAAACGTCTCACGACA................................................................................................................................. | 16 | 1 | 5 | 0.40 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................AAAGCACAAACGTCTCACGACA................................................................................................................................. | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................AGGTGGCGAGAGGTTG........ | 16 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................GCACAAACGTCTCACGACA................................................................................................................................. | 19 | 3 | 19 | 0.32 | 6 | 0 | 1 | 2 | 3 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................AAGCACAAACGTCTCACG.................................................................................................................................... | 18 | 2 | 7 | 0.29 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................................GTCAGGTGTCGAGAGGTTG........ | 19 | 3 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................AGCACAAACGTCTCACGACA................................................................................................................................. | 20 | 3 | 10 | 0.20 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................AAACGTCTCACGACA................................................................................................................................. | 15 | 1 | 15 | 0.20 | 3 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................GTCAGGTGGCGAGACGTTGA....... | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................TCAGGTGGCGAGAGGTTG........ | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................TCTCACGTCATGGTAGCCGC....................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................TGTCAGGTGGCGAGAGGTTG........ | 20 | 3 | 13 | 0.15 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................TAATGTCGCGAGAGG........... | 15 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................TAATGTTGCGAGAGG........... | 15 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................AGGTGTCGAGAGGTTGA....... | 17 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ........................................................................................................AGAAACGTCTCACGACA................................................................................................................................. | 17 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................GTCAGGTGGCGAGAGGTT......... | 18 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................AAGCACAAACGTCTCAC..................................................................................................................................... | 17 | 2 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................CAGGTGGCGAGAGGTTG........ | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................TACGTTGACAAGTCG..................................... | 15 | 1 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................AAAGCACAAACGTCTCAC..................................................................................................................................... | 18 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................GAAAGCACAAACGTCTCACG.................................................................................................................................... | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................GTGGAGAGAGCTTGAGATC... | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................CTTCCTTGTTTTATG................................................................................... | 15 | 1 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........TAACGTCGTCGCTGCATTTAGTT......................................................................................................................................................................................................................... | 23 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................AGCACAAACGTCTCACG.................................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................GGTGTCGAGAGGTTGA....... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................AGTGACGTTTGGAAAG....................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 2r:7214446-7214695 + | dsi_11697 | CA---GGCGGCAAATTG----------------------------------------------------------------------------CAGCAGCAGCGA-----------AA--ATCAAATATAAATAGCGAAAAGG-TAAG--G--AC----AGAGGCCCCAC------------A---AACT-------------------AAACTAAAAATAAGAG------------------------------CCGA-ACATA-TTTGTTGTTTGCAGA-----------------------------G----T-G-----CAGTAA-AATGGGCGC---AAATAAATGGGCA-----------------------------------AAAGAA----AGAAGCAA------CA-AAATACTGC----AAACCTTTCAATTGGCTTCAATTGGCAAATGCAAT-TGTTCAGCATTACA--TTTGCATTACACCGC------------------------------------------------------------------TCTCCA-------------------ACTCTTGCTC--- |
| droSec2 | scaffold_1:4057493-4057737 + | CA---AGCGGCAAATTG----------------------------------------------------------------------------CAGCAGCAGCGA-----------AA--ATCAAATATAAATAGCGAAAAGG-TAAG--G--AC----AGAGGCCCCAC------------A---AACT-------------------AAACTAA-AATAAGAG------------------------------CCGA-ACATA-TTTGTTGTTTGCGGA-----------------------------G----T-G-----CAGTAA-AATGGGGGC---AAATAAATGGGCA-----------------------------------AAAG-----AAGAAGCAA------CA-AAATACTGC----AAA-CTTTCAATTGGCTTCAATTGGCAAATGCAA--TGTTCAGCATTACA--TTTGCATTACACCGC------------------------------------------------------------------TCTC-A-------------------ACTCTTGCTC--- | |
| dm3 | chr2R:6445989-6446238 + | CA---GGCGGCAAATTG----------------------------------------------------------------------------CAGCAGCAGCGA-----------AA--ATCAAATATAAATAGCGAAAAGG-TAAG--G--AC----AGAGGCCCCAC------------A---AACT-------------------AAACTAAAAATAAGAG------------------------------CCGA-ACATA-TTTGTTGTTTGCAGA-----------------------------G----T-G-----CAGTAA-AATGGGTGC---AAATAAATGGGCA-----------------------------------AAAGAA----AGAAGCAA------CA-AAATACTGC----AAACCTTTCAATTGGCTTCAATTGGCAAATGTAAT-TGTTCAGCATTACA--TTTGCATTACACCGC------------------------------------------------------------------TCACCA-------------------ACTCTTGCTC--- | |
| droEre2 | scaffold_4845:19304398-19304647 - | CA---GGCGGCAAATTG----------------------------------------------------------------------------CAGCAGCAGCGA-----------AA--ATCAAATATAAATAGCGAAAAGG-TAAG--G--AC----AGCGGCCCCAC------------A---AACT-------------------CAACTAAAAATAAGAG------------------------------GCGAAACATA-TTTGTTGTTTGCAGA-----------------------------G----T-G-----CAATAA-AATGGGTGC---AAATAAATGGGCA-----------------------------------AAAG-----AAGAAGCTG------CA-AAATACTGC----AAACCTTTCAATTGGTTTCAATTGGCAAATGCAAT-TGTTCAGCGTTACA--TTTGCATTACACCGC------------------------------------------------------------------TCTCCC-------------------ACTCTCTCTC--- | |
| droYak3 | 2R:2693420-2693661 - | CA---GGCGGCAAATTG----------------------------------------------------------------------------CAGCAGCAGCGA-----------AA--ATCAAATATAAATAGCGAAAAGG-TAAG--G--AC----AGCGGCCCCAC------------A---AACT-------------------CAACTAAAAATAAGAG------------------------------GCAA-ACATA-TTTGTTGTTTGCAGA-----------------------------G----T-G-----CAATAA-AATGGGTGC---ATATAA---------------------------------------TATAAAG-----CAGAAGCTG------CA-AAATACTGC----AAACCTTTCAATTGGTTTCAATTGGCA-ATGCAAT-TGTTCAGCATTACA--TTTGCATTACACCGC------------------------------------------------------------------TCTCCA-------------------ACTCT--CTC--- | |
| droEug1 | scf7180000409672:4564233-4564477 - | CA---GGCGGCAAATTG----------------------------------------------------------------------------CAGCAGCAGCGA-----------AA--ATCAAATATAAATAGCGAAAAGG-TAAC--G--AC----AGA-GCCCCAC------------AA--AACT-------------------AAACTAAAAATAAGAG------------------------------GCGA-ACATA-TTTGTTGTTTGCTGA-----------------------------G----T-G-----CAATAA-AAAGGATG-------------G-CA----A--A--GCAGAGG--GGAGAGAATAAAATTTAAA---------------AAATT-CA-AAATACTGC----AAACCTTTCAATTGGTTTCAATTGGCAAATGCAAT-TGTTCAGCATTACA--TTTGCATTACATCGC------------------------------------------------------------------TCT----------------------------------- | |
| droBia1 | scf7180000301506:153700-153932 + | CA---GGCGTCAAATTG----------------------------------------------------------------------------CAGCAGCAGCGA-----------AA--ATCAAATATAAATAGCGAAAAGG-TACA--GAGGCTGA-GCA-GCCCCGC------------GA--AACT-------------------CAACTAAAAATAAGAG------------------------------GCGA-ACATA-TTTGTTGTTTGCACA-----------------------------G----T-G-----CAATA----------C---AAAAAAATGG-----TGG--GGGG--GAGGG------------------------------------G------GGTATACTGT----AAACCTTTCAATTGGCTTCAATTGGCAAATGCAAT-TGTTCAGCGTTACA--TTTGCATTACATCGC------------------------------------------------------------------GCTC---------------------------CCTC--- | |
| droTak1 | scf7180000415877:381697-381926 - | CA---GGCGGCAAATTG----------------------------------------------------------------------------CAGCAGCAGCGA-----------AA--ATCAAATATAAATAGCGAAAAGG-TAAA--GAGAC----AGA-GTCCCAC------------AA--AACC-------------------AAACTAAAAATAAGAG------------------------------GCGA-ACATA-TTTGTTGTTTGCGGA-----------------------------G----T-G-----CA-TAAAAAAAGAG-------------GG-------------G--CAGGGGAGA---A---------------------------AAAATA--AAAATACTGC----AAACCTTTCAATTGGTTTCAATTGGCAAATGCAAT-TGTTCAGCATTACA--TTTGCATTACATCGC------------------------------------------------------------------TCTC------------------------------C--- | |
| droEle1 | scf7180000491349:1005836-1006034 - | CA---GGCGGCAAATTG----------------------------------------------------------------------------CAGCAGCAGCGA-----------AA--ATCAAATATAAATAGCGAAAAGG-TAAA--G--GC----GCA-GCCCCAC------------AA--AACT-------------------AAACTAAAAATAAGAG------------------------------ACGA-ACATA-TTTGTAGTTTGAGGG-----------------------------G----A--------------------------------------------------------------------------------------------AAAATA--AAAATACTGC----AAACCTTTCAATTGGTTTCAATTGGCAAATGCAAT-TGTTCTGCATTACA--TTTACATTACAGCGC------------------------------------------------------------------TCT----------------------------------- | |
| droRho1 | scf7180000779999:312809-313023 - | CA---GGCGGCAAATTG----------------------------------------------------------------------------CAGCAGCAGCGA-----------AA--ATCAAATATAAATAGCGAAAAGG-TAAA--G--GC----GCA-GCCCCAC------------AA--AACT-------------------AAACTAAAAATAAGAG------------------------------GCGA-ACATA-TTTGTTGTTTGAGGG-----------------------------G---------------------------------------------------------------GAA---A---------------------------AAAATA--AAAATACTGC----AAACCTTTCAATTGGTTTCAATTGGCAAATGCAAT-TGTTCAGCATTACA--TTTGCATTACAGCGC------------------------------------------------------------------TCTCTT-------------------TCTCTCTCTC--- | |
| droFic1 | scf7180000454066:1234769-1234991 - | CA---GGCGGCAAATTG----------------------------------------------------------------------------CAGCAGCAGCGA-----------AA--ATCAAATATAAATAGCGAAAAGG-TAAA--G--GC----A---GCCCCAC------------AAA-AACT-------------------AAACTAAAAATAAGAG------------------------------GCGA-ACATA-TTTGTTGTTTGCGAA-----------------------------G----T-G-----TA-TAA-AAAGGAG-------------------------G--G-----G--GGAGGGA---------------------------AAAATA--AAAATACTGC----AAACCTTTCAATTGGTTTCAATTGGCAAATGCAAT-TGTTCAGCATTACA--TTTGCATTACATCGC------------------------------------------------------------------TCTC------------------------------C--- | |
| droKik1 | scf7180000302406:173316-173562 + | CA---GGCGGCAAATTG----------------------------------------------------------------------------CAGCAGCAG--------------AA--ATTCAGTATAAATAGA-AAAAGG-TAAA--AAAAAA--CAAGAA-AACAAGA---CGA--------GCGTGCACTA-------------AAAATAAAAATAAGAG---------------------------------------------TTGTTTGAAAA---------------------------A-G----T-GATCTACAATA-----------CAG-------------AAAAAAGG--G--GG---------------------------------------GAATA--AAAATACTGC----AAACCTTTCAATTGGTTTCAATTGGCAAATGCAAT-TGTTCAGCATTACA--TTTGCATTACATTGC-------------------------------------ATT--GGGGTTGCAC--C-----------TATCCC-------------------GCTCCCACTC--- | |
| droAna3 | scaffold_13266:17936643-17936919 + | CG---GGCGGCAAACTG----------------------------------------------------------------------------CAGCAGCACC--------------A--TACAAATATAAATAGCGGAAAGG-TAAA--AAAA------------------------------AAAACA-----------AAAGCAA-CAACT----ATAGGAG------------------------------GGGG-AGGAATTT---TGTTTTAAAGGTTTAAAGGGGTCTAGGGGGGA--------------TATCT---------------AC---AAATAAA------A-------------------------------AATAAGG----C----------AAAAA--AAAATACTGCTGAAAAACCTTTCAATTGGTTTCAATTGGCAAATGCAAT-TGTTCAGCATTACA--TTTGCATTACATTGC-CCGCGTGGGAGGGGGGGACAG-----------AA--TAAGG----------GGGACTG-CAGCGCAC------------------------------CCC--- | |
| droBip1 | scf7180000396735:1192543-1192811 + | CG---GGCGGCAAACTG----------------------------------------------------------------------------CAGCAGCACC--------------A--CTCAAATATAAATAGCGAAAAGG-TAAA--G------------------C------------AA--AAGC----------------AA-CAACTAAAAATAAGAG------------------------------CGGT-------TT---AGTTTGCG--GTTTG--GGGGTTTGGGGGGGACCTAAACTTGGATATATCT---------------AC---AAATAA-------------------------------------------------------------------A-AAATACTGCTGA-AAACCTTTCAATTGGTTTCAATTGGCAAATGCAAT-TGTTCAGCATTACA--TTTGCATTACATTGC-CAG----AGAGGGGAGTGCAG-----------CA--GGGGG----------GGGAGTG------CACC-------------CCACTTGC-ACTTTTGCTG--- | |
| dp5 | 3:16161255-16161607 - | CA-------ACAAATTGAACGCTGGCGGCAGCGATGAGCTCGTCCCACCCATTGCAGCAATACCAGTAGCAGAATCCGCAGCAATAGCAGCAGCAGCAGCAGCGAAATAAGCGCAAAACGCAAAAATATAAATAGCGAAAAGG-TAAGTCG--GC----A---GC---AG------------AAACAACA-------------------AAACTAAAAATAAGAGTTGGCGCTCGCCAAAGCGGGCAGTGCGCAGGCCG-AGATA-TTTGTG-------AA---------------------------ACG----T-AACGAAAAAAA-----------CTG-------------AATGAAGA--G--GA---------------------------------------G--------------------GACAATTTCAATTGGATTCAATTG-CAAATAAAATTTGTTCAGCATTACA--TTTACAT-ACAGTCC-------------------------------------GAGGG----------GGCACTG-TGACGCC-C-------------CCAGCCGCAGCCCCCGTCC--- | |
| droPer2 | scaffold_4:603360-603702 + | CA-------ACAAATTGAACGCTGGCGGCAGCGATGAGCTCGTCCCACCCATTGCAGCAATACCAGTAGCAGAATCCGCA---------GCAGCAGCAGCAGCGAAATAAGCGCAAAACGCAAAAATATAAATAGCGAAAAGG-TAAGTCG--GC----A---GC---AG------------AAACAACA-------------------AAACTAAAAATAAGAGTTGGCGCTCGCCAAAGCGGGCAGTGCGCAGGCCG-AGATA-TTTGTG-------AA---------------------------ACG----T-AACG-AAAAAA-----------CTG-------------AATGAAGA--G--GA---------------------------------------G--------------------GACAATTTCAATTGGATTCAATTG-CAAATAAAATTTGTTCAGCATTACA--TTTACAT-ACACTCC-------------------------------------GAGGG----------GGCACTG-TGACGCC-C-------------CCAGCCGCAGCCCCCGTCC--- | |
| droWil2 | scf2_1100000004516:1241000-1241064 + | CTTCAGTAGGCAGCCTG----------------------------------------------------------------------------GA-----------------------------------------------------------------CA-G------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTGGTTTGACTTGAC------------------------------TACTTTACCCC------------------------------------------------------------------TCTCTA-------------------TCTCTCTCTC--- | |
| droVir3 | scaffold_12875:16431594-16431858 + | T--------------TG----------------------------------------------------------------------------CAGCGAC----A-----------AA--TTTAAATATAAATAGCAACAACAACAAC--A--AC----AACAA-CACACGAAAAGGTAAAC------------CAACAATAACGCAACAAACTAAAAATAATAA-------------------------------------------------TAGCAAC-----------------------------G---GT-A-----C------------------AAAAAA------------------------------------------AAAA-----CACAAACAC------AC-AAATTTAAC----ACACGTTTCAATTGATTTCAATTG-CGACTA-----TATTAAGCATTACA--TTTAT-TTACATGTGGCGG----AAGTGAGCGTGCGG-----GGGAGG--GG----AG--------GGGGTATG-CC-----GTGCACATTTCCATGCC-------ACCTGCACTTCGG | |
| droMoj3 | scaffold_6496:7331314-7331531 - | CA-------ACAAATTG----------------------------------------------------------------------------CAA-AGC-TCTA-----------AA--TTTAAATAGAAATAGCAACAA-----------------------CACCAA----AGGT--------AAAT-------------------CAACTAAAAATAAA-G------------------------------GCAA-GCAAA-----------------------------------------------------AAGAA---------------------------------------------------GAA---A---------------------------AAAAAA--AACAGAAGCA----ACACATTTCAATTGATGTCAATTGC----GGCAA--TGTTAAGCATTACACATTTAT-TTACATGGG-CAA----A--------TATAGTGGGGGGC-------ATA--CGACTTATTC--GTATA-TC-----GTGCA-------------------AATTTCCATA--- | |
| droGri2 | scaffold_15245:5278778-5278998 + | TTGCAACAGCCAAATTC----------------------------------------------------------------------------AAAGC---GCTA-----------AA--TTTATATATAAATAGCAACAACAACAAA--A--AA-------------------AGGTAAAC------------TACAAACAAAGCTA-CAACTAAAAATAAGAG------------------------------A--A-AC------------------------------------------------GTCGAT-T-----C------------------AAG----------------------------------------------AAGAAAGC---------AAAAAA--AAAATACT---------CATTTCAATTGGTTTCAATTGCGA----CAA--CGTTGAGCATTACA--TTTAT-TTACATGTGGCGG-------------------------------GG----CA--------G--GGATGTC-----TGTGCA-------------------AATTTCCATG--- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 10:12 AM