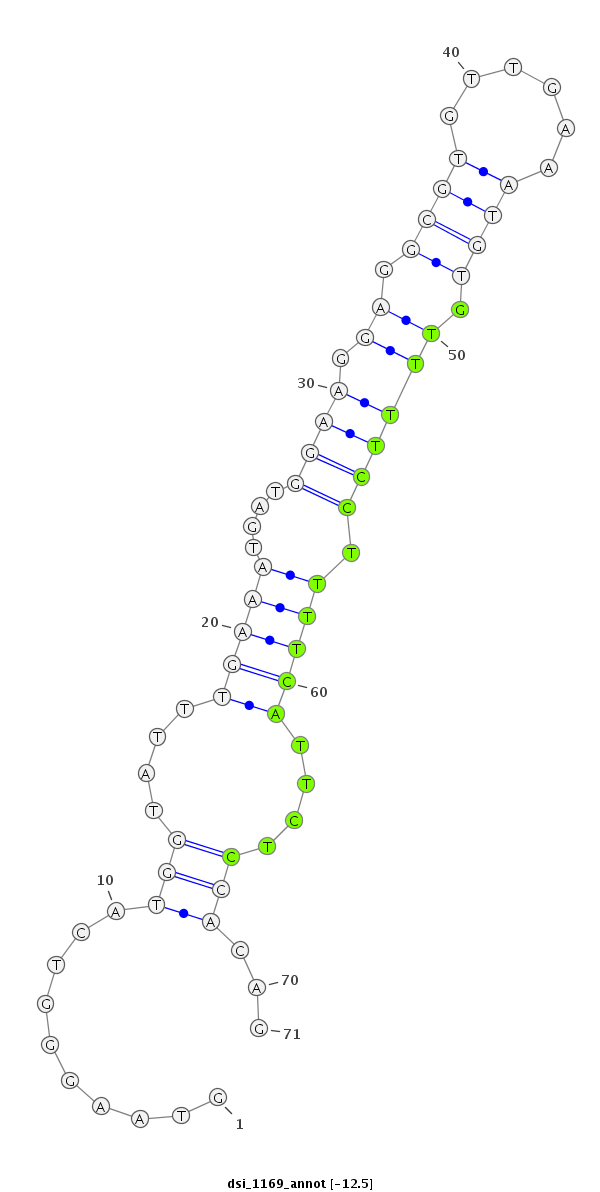
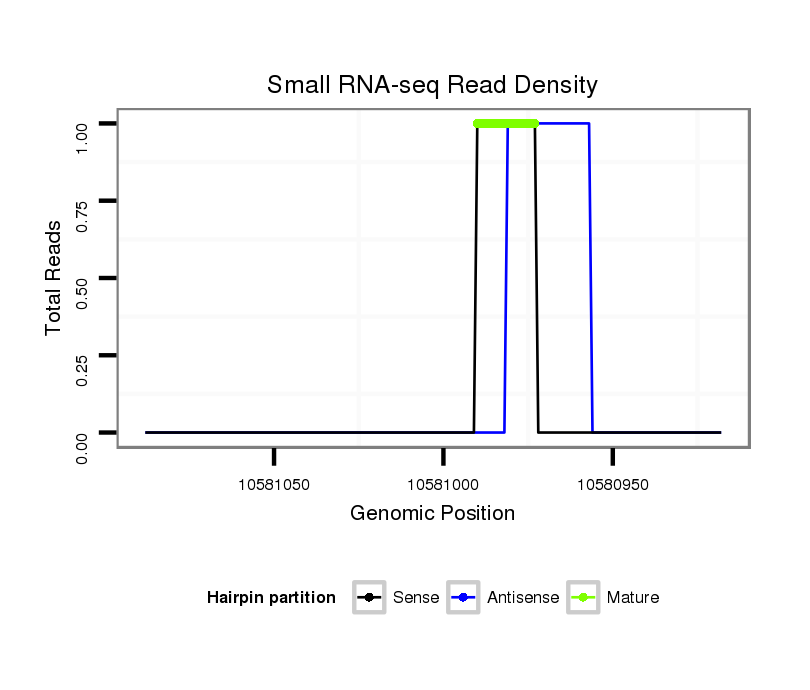
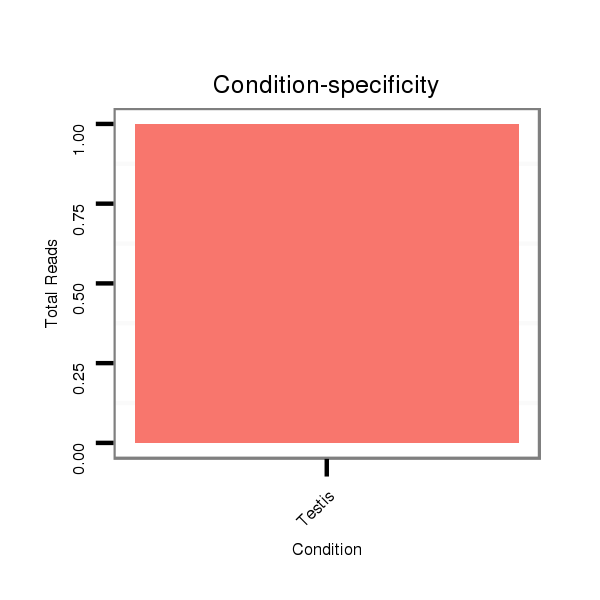
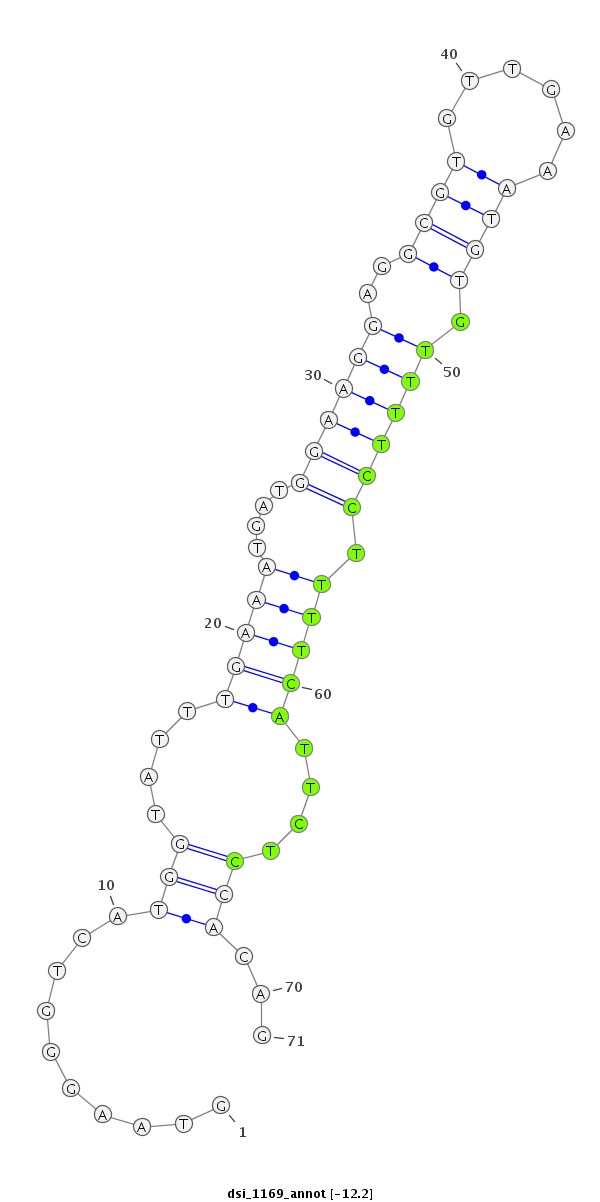
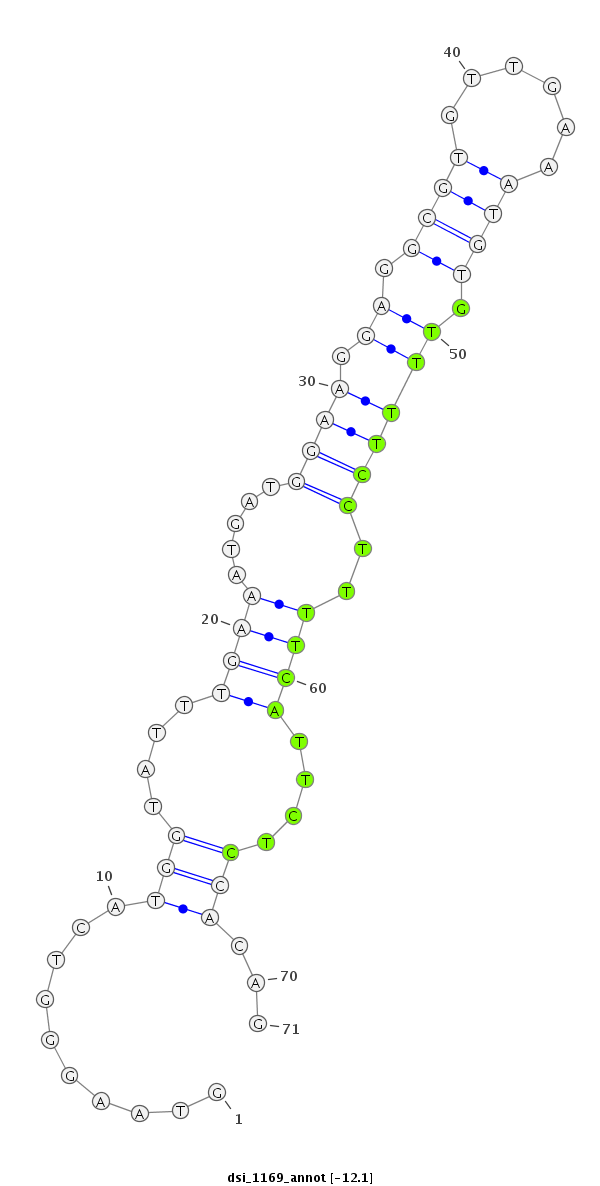
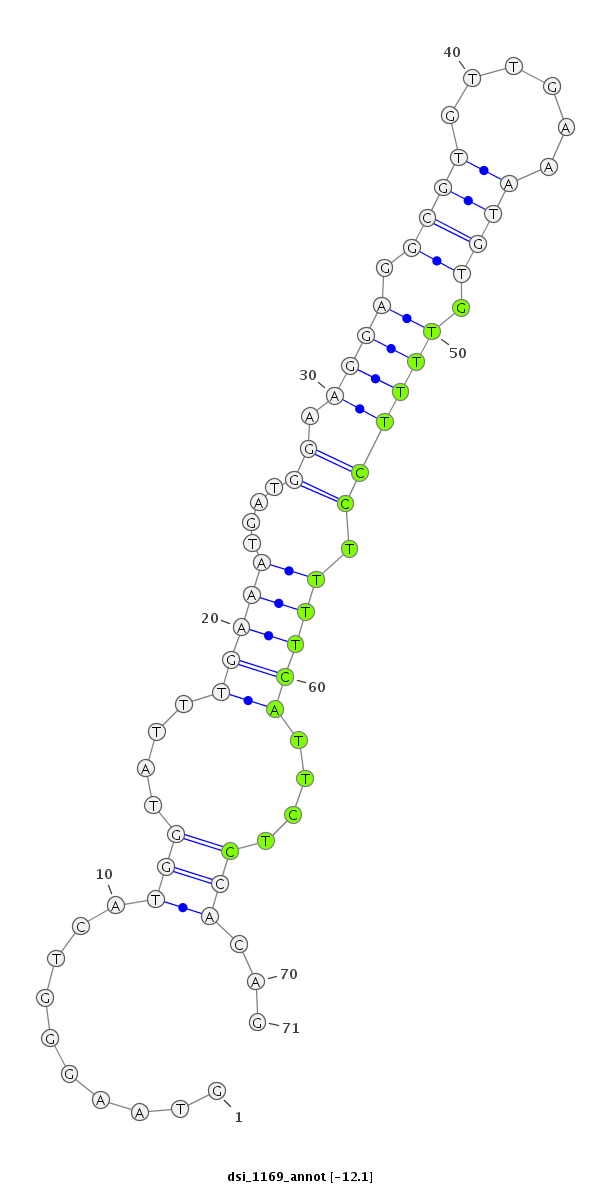
ID:dsi_1169 |
Coordinate:3r:10580968-10581038 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -12.2 | -12.1 | -12.1 |
 |
 |
 |
CDS [3r_10580796_10580967_-]; exon [3r_10580438_10580967_-]; CDS [3r_10581039_10581545_-]; exon [3r_10581039_10581545_-]; intron [3r_10580968_10581038_-]
No Repeatable elements found
| ##################################################-----------------------------------------------------------------------################################################## CTACTTACCGCATCCGGATGCACTCTCATGCTGCCGGAAACGAACAATAGGTAAGGGTCATGGTATTTGAAATGATGGAAGGAGGCGTGTTGAAATGTGTTTTCCTTTTCATTCTCCACAGACAACTGCCCATCACATTGGCTGATGGAGAGGCATTTGGGAAGGGCGAAT **************************************************..........(((....(((((....((((.((.((((......)))).)))))).)))))....)))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
SRR618934 dsim w501 ovaries |
SRR1275487 Male larvae |
SRR902009 testis |
SRR553488 RT_0-2 hours eggs |
SRR1275483 Male prepupae |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................GTTTTCCTTTTCATTCTC....................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................GAAGGAGGCGTGGTGA.............................................................................. | 16 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................AGAAGGCGTGCTGGAATGTG........................................................................ | 20 | 3 | 9 | 0.44 | 4 | 0 | 0 | 2 | 2 | 0 | 0 | 0 |
| .........................................................................................................TTTTCCTTCTCCACAGA................................................. | 17 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................AGGCGTGCTGGAATGTG........................................................................ | 17 | 2 | 19 | 0.11 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| ..............................CTGCCGGAACCGACCAA............................................................................................................................ | 17 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....TACCGAATCCTGATGAACT................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................ACTATAGGTAAGGGC................................................................................................................. | 15 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
|
GATGAATGGCGTAGGCCTACGTGAGAGTACGACGGCCTTTGCTTGTTATCCATTCCCAGTACCATAAACTTTACTACCTTCCTCCGCACAACTTTACACAAAAGGAAAAGTAAGAGGTGTCTGTTGACGGGTAGTGTAACCGACTACCTCTCCGTAAACCCTTCCCGCTTA
**************************************************..........(((....(((((....((((.((.((((......)))).)))))).)))))....)))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
M023 head |
SRR902009 testis |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
M053 female body |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................AAGTAAGAGGTGTCTGTTGACGGGT....................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................ACAGAAGGAAAAGTAGGAGGCG.................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................GACGGCCTTTGCTTCTTC........................................................................................................................... | 18 | 2 | 16 | 0.13 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TCTTATCCATTTCCAGTA.............................................................................................................. | 18 | 2 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................AGTGTAACCTACTAC........................ | 15 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................AAAGGAAGAGGTGTCT................................................. | 16 | 1 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................TCTGTTGACTGGTACTG................................... | 17 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................CCCTCCGCACGACTTGACA......................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................GATGGCCTATGCTTG.............................................................................................................................. | 15 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................CATCCGGGCAACTTTACAC........................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................................GTCTGTTGAAGGGTATT.................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
Generated: 04/24/2015 at 10:45 AM