| droSim2 |
2l:2129827-2130076 - |
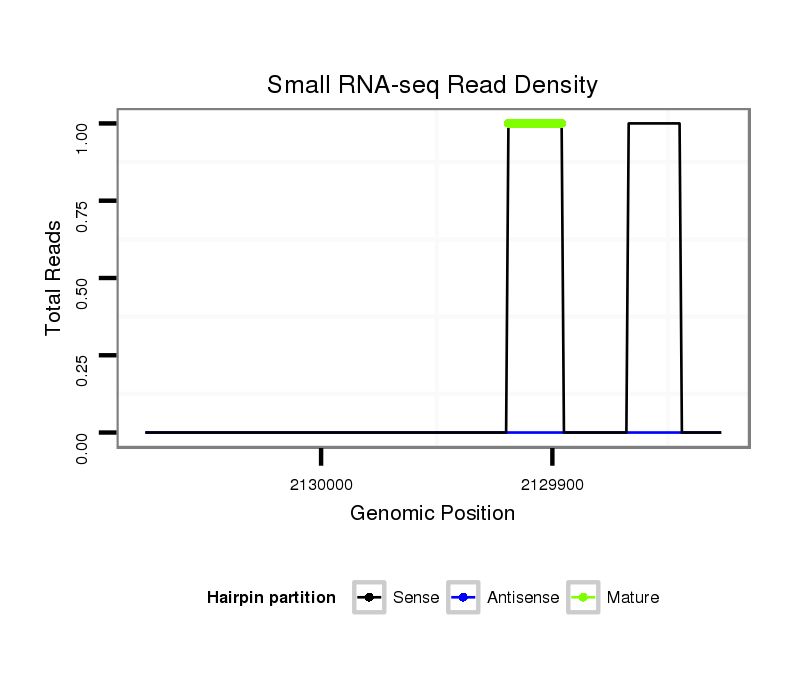
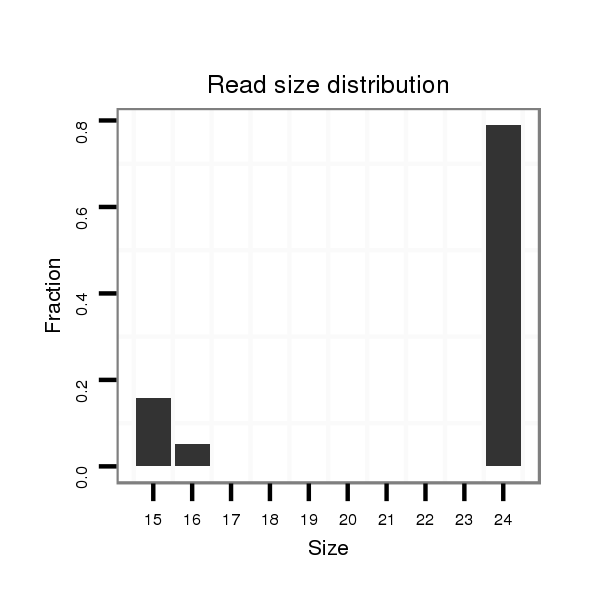
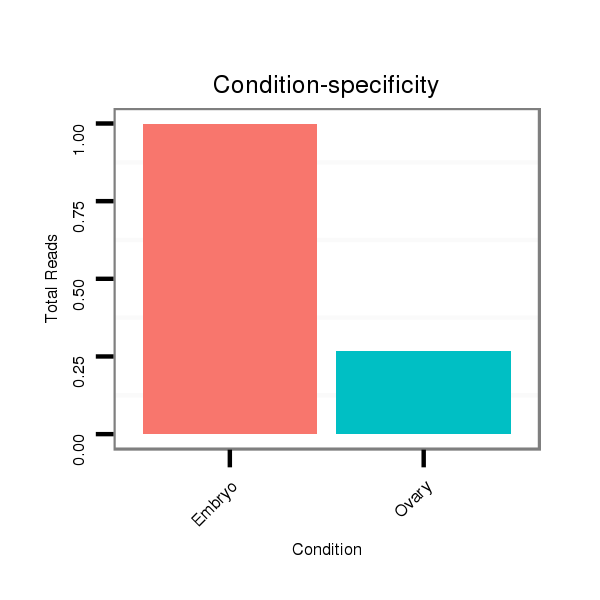
dsi_11669 |
CAAC--------------GTCGGCGGC---------------A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCCCAGCCTTACCTCAATGGCAACTACCAGCAGCAGGTGAATAG-ATAGCTTTAACCATGGCAGCTTTAACAAGAATTGGAAAACAATCCCG---------------------CTGCACTCGTCAAATATATGTGTATTC-----CCTTCAACTT------CATTCTGGC-------------------------------GATAGCCTATT------G---GG---------------C---------------------------------TAGTTGATAATAAGAATAGGGTTAGAATCTGAAATGAATTGCAGAATTACGTTACTCTTTTGAACCAATTGGCCACCTTGATAT------------------------------- |
| droSec2 |
scaffold_5:411095-411338 - |
|
CAAC--------------GTCGGCGGC---------------A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCCCAGCCTTACCTCAATGGCAACTACCAGCAGCAGGTGAATAG-ATAGCTTTAACCATGGCAGCTTTAACAAGAATTGGAAGACAATCCCG---------------------CTGCACACGTCAAATATATGTGTATTC-----CCTCCGGCTT------CATTCTGGC-------------------------------AATAGACTATT------G---GG---------------C---------------------------------TAGTTGATAATAAGAATAGGGTTAGAATCTGAAATGAATTGCAGAATTACGTTACTCTTTTGAACCAATTGGCCACCT------------------------------------- |
| dm3 |
chr2L:2236108-2236357 - |
|
CAAC--------------GTCGGCGGC---------------A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCCCAGCCTTACCTCAATGGCAACTACCAGCAGCAGGTGAATAG-ATAGCCTTAACCATGGCAGCTTTGACGAGAATCGGAAGACAATCCGG---------------------TTGCACACGTCAAATATATGTGTATTC-----CCTTCAGCTT------CATTCTGGC-------------------------------AATAGCCTATT------G---AG---------------C---------------------------------TAGTCGATAATAAGAATAGGGTTAGAATCTGAAAAGCATTGCAGAATTACGTTACTCTTTTGAACCAATTGGCCACCTTGATAT------------------------------- |
| droEre2 |
scaffold_4929:2271442-2271723 - |
|
CAAC--------------GTCGGCAGC---------------A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCCCAGCCTTACCTCAATGGCAACTACCAACAGCAGGTGAATGGGATAGCTTTATGCATGATAGCCTTAACAAGAATTTGCAGACCATCC-----------------------------------------TGTATAATC-----CGCCCAGCTT------CATCCTGGTCTTCTTGATTTTCACATTTGGATCAAACTGAAGTAGCCCATC------A---AT----------------TTTCATCACAGAATTTATAAATATTATG-------ATTGACAATAAGAATAGGGTTAGAATCTGAAAG------------------------------CCATTGGT---ATTGATATAATTA---ACATACATATTTAAAACTAGTTT |
| droYak3 |
2L:2209203-2209462 - |
|
CAAC--------------GTCGGCGGC---------------A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCCCAGCCATACCTCAATGGCAACTACCAGCAGCAGGTGAATGGCATAGCTTTATCCATGATAGCTTTAAT-AGAATGGACGAAGCATCC-----------------------------------------TGTAAATTC-----CGGCCAGCTT------CATCCTGGTCTTCTTCAAT--------------AAACTGAAATAGTCCATC------A---AG---------------CTTTTATTATTGAATTTATCAATATTATGATCCCTAATTGACAATAAGAAAAGAGTTAGAATCTGAA-------------------------------TCCATTGGT---ATTGATATAATTAGAAATATAC----------------- |
| droEug1 |
scf7180000409711:4775247-4775398 - |
|
CCGC--------------AGCAGCAAT---------------C------TTTTGGCCTAGCGGACAGCAGCAAC---AACAA---CAACAACAACAACAG------------------------------------CAACGGCAGCAGCA------------GCAACAACAACAACA---GCAA----------------C--------------------------------------------------------------------------------------------------------------------------------GGCGTCTCCTCGAA-----A-------TCATTTGTG----C-----CCTGCAAAGTCTG--------TGGC-------------------------------GACAAGGCATC------G---GG---------------C---------------------------------TA----------------------------------------------------------------------------------------------------------------- |
| droBia1 |
scf7180000302193:4804497-4804642 - |
|
CCGC--------------AGCAGCAAT---------------C------TTTTGGCCTAGCCGACAGCAGCAAC---AACAACAGCAACGGC---AG---------------------------------------------------CAACAGCAACA---GCAACAGCAACAACA---CCAA----------------C--------------------------------------------------------------------------------------------------------------------------------GGGGTCTCCTCGAA-----G-------TCATTTGTG----C-----CCTGCAAAGTCTG--------TGGC-------------------------------GACAAGGCATC------G---GG---------------C---------------------------------TA----------------------------------------------------------------------------------------------------------------- |
| droTak1 |
scf7180000415310:699302-699467 - |
|
CAAC--------------CGCAGCAGC---------------AG-----------------------C---AACAAGACCAA---CAGCAACAGCAACAACAACATGTGTTGCAA---G------------------------TGCAGCA---ACCGCAGCAGCAACAACCG------------------------TTAA----------------------------------------------------------------------------------------------------------TGCAG-----------CCG---CAGTCGCCCCATCC---------A----TCAAGTGCGTTTTCTTTTCCCTACACTTT------CACCGAAAC-------------------------------AATCTCTTATT------A---AA---------------C---------------------------------CA----------------------------------------------------------------------------------------------------------------- |
| droEle1 |
scf7180000491338:149046-149236 - |
|
CAAT--------------CGCTTCAAC---------------AG-----------------------C---AACAA---CAC---------------------CATCCGGTCTACAGCAACTTTCAG---------CAAC---AGCAGCA------------GCAGCAACAACAACA------------ACAG---------------------------------------------------------------------------------------------------------------------------------GCTGTCCCTGGTCTCAATGAG-------------CCTCGTGTGCCTCC-----CCTGCAAATCCGTTTGCGCGGCA------------------------------------AG------AACCATGTGG--------------------------------------------TG-------GTC------AAGAATACGCGAAAAGATCGAAAGAAGGGACAGAATCA------------------------------------------------------------------ |
| droRho1 |
scf7180000777865:82788-82945 + |
|
CAGC--------------AAGTGCAGC---------------AA-----------------------C---TCCATCAACCG---CAACAGCAGCAGCAA---CATGTGCTGCAA---------------------------------AC---GCAACA---GCCACAACAGCAACA------------ACCG---TTAA---------------------------------------------------------------------------------------------------------------------TGCAGCCG---CAGTCACCCCATCC-------------ATCAAGTGAGTTTTT-----CATATACTTT------CACCAAAAC-------------------------------AATCGCTTTTT------A---AA---------------C---------------------------------CA----------------------------------------------------------------------------------------------------------------- |
| droFic1 |
scf7180000453929:375769-375926 + |
|
CCGC--------------AGCAGCAAT---------------C------TTTCGGCCTCGCCGACAGCAGCAAC---AGCAACGGCAGCAGC---AG------------------------------CAACAACAACAACAACTGCAGCA------------ACAGCAACAACAACA---GCAA----------------C--------------------------------------------------------------------------------------------------------------------------------GGAGTCTCCTCGAA-----G-------TCATTTGTG----C-----CCTGCAAAGTCTG--------CGGC-------------------------------GACAAGGCATC------G---GG---------------C---------------------------------TA----------------------------------------------------------------------------------------------------------------- |
| droKik1 |
scf7180000302577:465648-465806 + |
|
CAGC--------------AACACCAGC---------------AA-----------------------C---TGCTGCAGCAA---CAGCTGCCGCAGCAG---CG-------TAGACAAACCTACAAG--------GAATATTTACAGCA------------AAAGCAACAGCAACAGCAAAACAAGCAACCA---CTAAGGATAT-------------------------------------------------------------------------------------------------------------------------------------------------------TCCGCGTCCACTA-----CATTCGACTC------AACTCCA------------------------------------AGGACGGC------G---TA---------------C---------------------------------CG----------------------------------------------------------------------------------------------------------------- |
| droAna3 |
scaffold_12916:6235341-6235532 + |
|
CAATTTCCGGTGGATGGCCACCTCAACAGCAACCACTACAA--------------------------C---AGCAGCAGCAA---------------------CAGCCGTTGCCG------CCGCAGC---------------AGCAGCA------------GCAGCACCTGCAACA------------ACAACCAGCAC-AGTATGGAGCACCGCCGACGTCGTCAGCTCCCCAGACCTACCTGAATGGCAACTACCAACAGCAGGTGAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATCACGGGGTTCCAATC-------------------------------------------------------------------------------------- |
| droBip1 |
scf7180000396568:593923-594155 - |
|
CGATTTCCGGTGGATGGCCACCTCAACAGCAACCACTACAA--------------------------C---AGCAG---CAA---------------------CAGCCGTTGCCG------------------------------CCGCA------------GCAGCACCTGCAACA------------ACAACCAGCAC-AGTATGGAGCACCGCCGACGTCGTCAGCTCCCCAGACTTACCTGAATGGCAACTACCAACAGCAGGTGAA-------------ATCACGGGAATTTTAA---------------------------------------------------------------------------------------------TCT----------------------------------------------------------------------------------------------------------------------------------------TGACCTACAATACTGATTTACTAGACTTTTTTTTGGAATTCCTCCCCTTTG----------------------------------- |
| dp5 |
4_group3:7142108-7142309 - |
|
CAAC--------------ATCAACAGC---------------AG-----------------------C---AGCCACAAACA---CATCTGCAGCAACAT---CATCCGGTGTACACAAACTTTCAAC---------------AGCAGCA------------GCAACAACAGCAACA------------GCAA---------------------------------------------------------------------------------------------------------------------------------TCTGTACAAGGTCTCAACGAA-------------CCACGTGTGCCTCC-----CCTGCAGATTCGTTTGCGTGGAA------------------------------------AG------AATCATGTTG--------------------------------------------TA-------GTTAAGAATACGCGCAAGGA------TCGCAAGAAGTTGCAGA---------------------------------------------------------------------- |
| droPer2 |
scaffold_1:8589347-8589548 - |
|
CAAC--------------ATCAACAGC---------------AG-----------------------C---AGCCACAAACA---CATCTGCAGCAACAT---CATCCGGTGTACACAAACTTTCAAC---------------AGCAGCA------------GCAACAACAGCAACA------------GCAA---------------------------------------------------------------------------------------------------------------------------------TCTGTACAAGGTCTCAACGAA-------------CCACGTGTGCCTCC-----CCTGCAGATTCGTTTGCGTGGAA------------------------------------AG------AATCATGTTG--------------------------------------------TA-------GTTAAGAATACGCGCAAGGA------TCGCAAGAAGTTGCAGA---------------------------------------------------------------------- |
| droWil2 |
scf2_1100000004511:5776953-5777104 - |
|
CAAC--------------AACTGCAAC---------------AACAAGCATTTAGCTTAGCCGATTCCAGCAAC---AACAACAACAACAGC---AG---------------------------------------------------CAGCAACTCTA---ATAACAGCAACAACG---GCAA----------------C--------------------------------------------------------------------------------------------------------------------------------GGCGTCTCATCAAA-----A-------TCTTTTGTA----C-----CCTGCAAAGTTTG--------CGGT-------------------------------GACAAGGCATC------G---GG---------------C---------------------------------TA----------------------------------------------------------------------------------------------------------------- |
| droVir3 |
scaffold_12963:13664416-13664604 - |
|
CAAC--------------AACAGCAGC---------------CT-----------------------C---AGCAACAACAA---CAACAACAGCAG---------------------------------------------------CC------------TCAGCAACAACAACA-------------------------ATATGGCGCTGCGGTGACG---TCCGCTCAGCAGCCCTATGTGAATGGCAACTACCA---GCAGGTGA----------------------------------------------------------------------------------------------GCTTCT-----GAGTCTGATTTTTAAGCACAACG------------------------------------GA------GAACATGTTA--------------------------------------------TA-------GC----------------------------------------------ATCCCAATTTCGAATCG--------CTTCGTTGC------------------------------- |
| droMoj3 |
scaffold_6328:2997605-2997741 + |
|
CAGC--------------AGCAGCAGC---------------AG-----------------------C---AACAA---CAACTGCAAATGCAATTG---------------------------------------------------CA------------TCAACAACTGCAGCA------------GCAGTTAGCAT----------------------------------------------------------------------------------------------------------TTCAAATGCAATCC----------------CAACTGGCTACGCCAGTGCAGTCAATGAAGGTTC-----CGTCC-ACTT------CGCTG---C-------------------------------CACAGCC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droGri2 |
scaffold_15126:6199603-6199807 + |
|
AACA--------------GCCGCCGCC----------GCAGCAG-----------------------CAGCAACAA---CAACAACCGCCGC---AG------------------------------CAGCACCAACAACAACCGCAGCA------------GCAACCACCACAACA---GCAA----------------CAATATGGCGCTCCAACAACG---TCCGCTCAGCAGCCTTTTGTCAATGGCAACTATC---AGCAGGTAAA-------------------------------------------------------------------------------------------------------------------------------AC-------------------------------AGCAAAACATT------G---TGCAATACTAAATTAAAA---------------------------------CA-----------------------------------------ACT----TAA-----------------GCTTACTCTAGAAA------------------------------- |