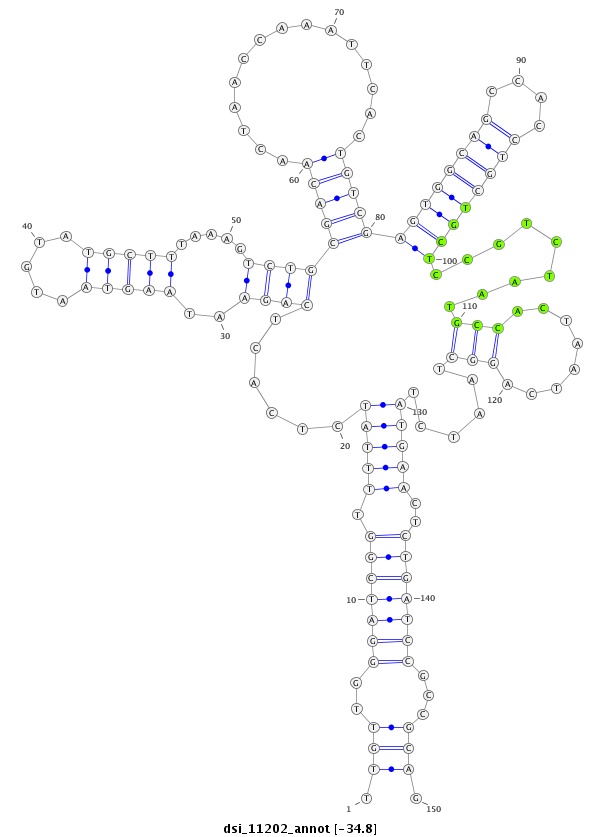
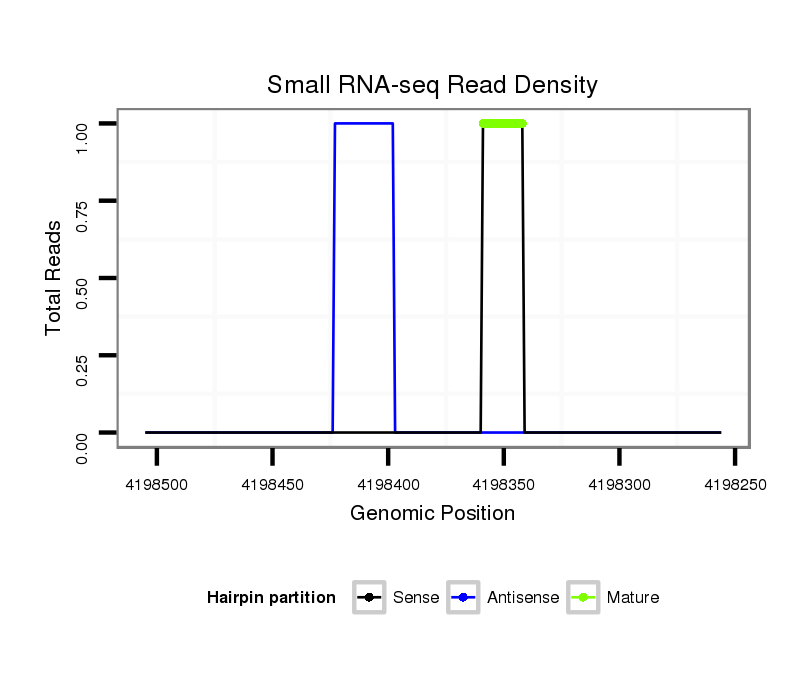
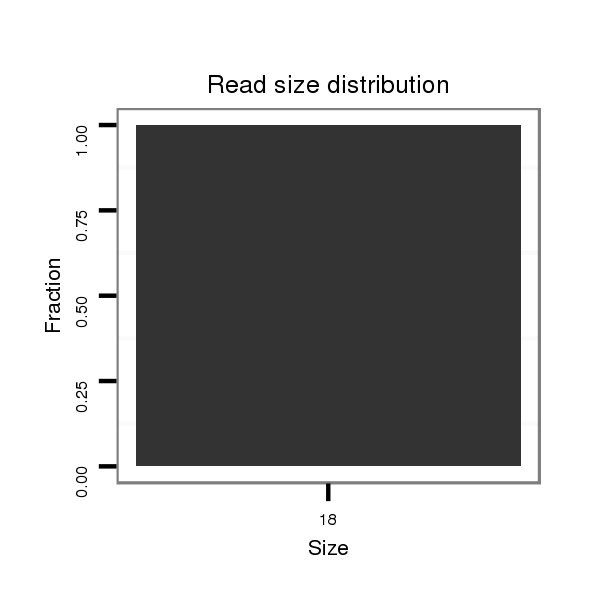
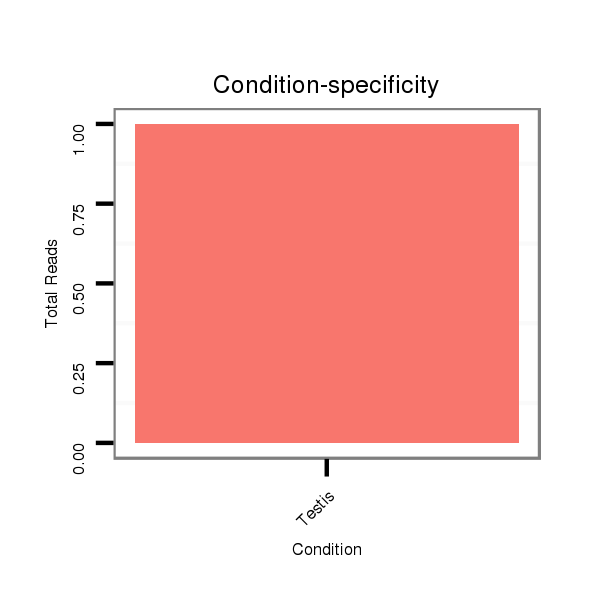
ID:dsi_11202 |
Coordinate:3r:4198306-4198455 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
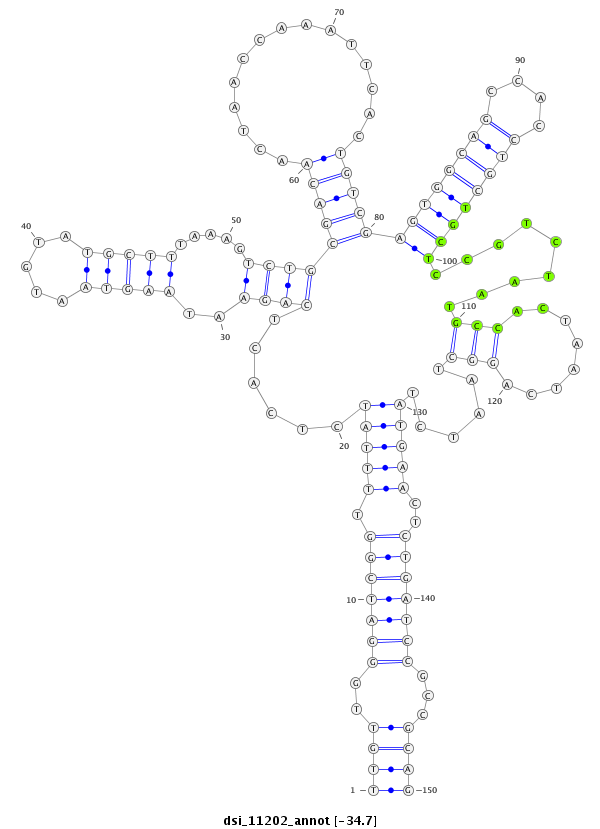
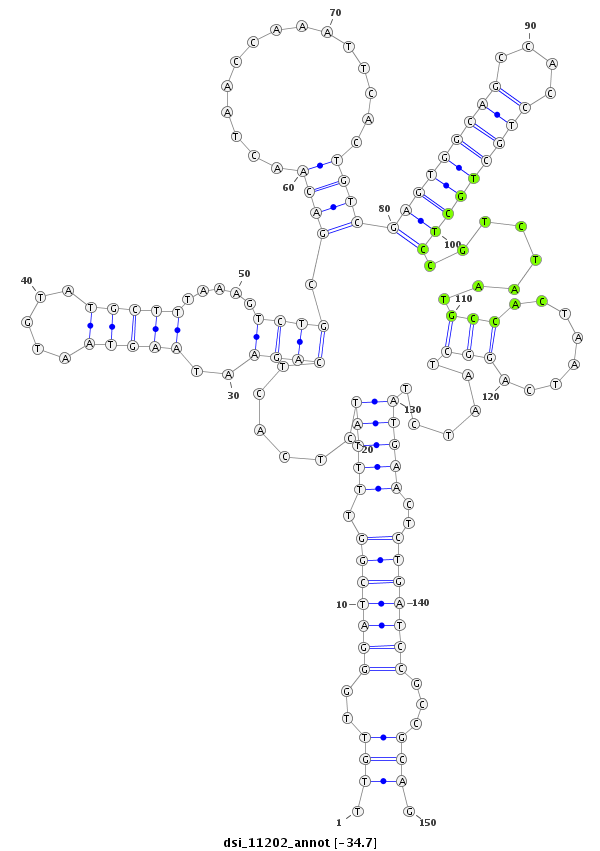
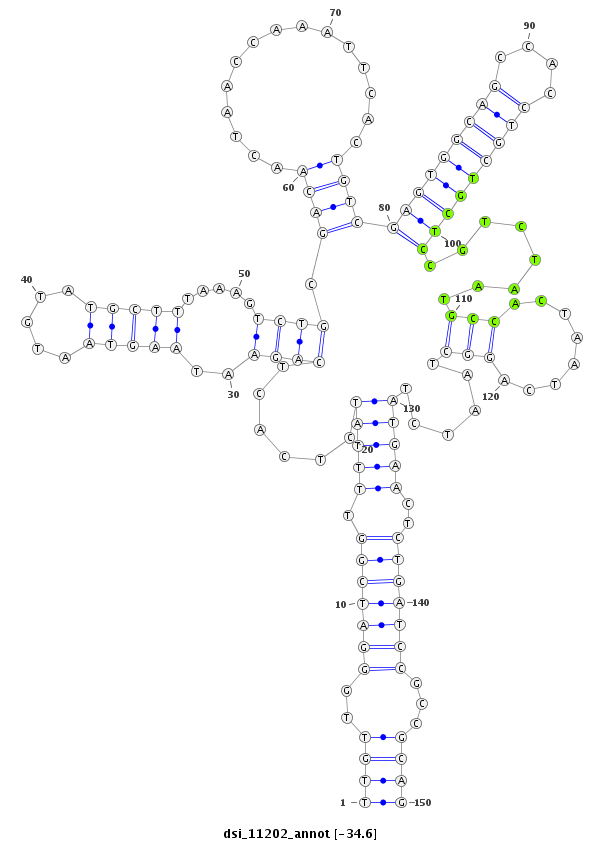
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -34.7 | -34.7 | -34.6 |
 |
 |
 |
CDS [3r_4198172_4198305_-]; exon [3r_4198172_4198305_-]; intron [3r_4198306_4198612_-]; Antisense to intron [3r_4197814_4201350_+]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ATTAAGTCTATATTACAACCCATTGAAGCTCATATGAGTCACCTAGTATGTTGTTGGGATCGGTTTTATCTCACTCAGAATAAGTAATGTATGCTTTAAAGTCTGCGACAACTAACCAAATTCACTGTCGAGTGGCAGCCACCTGCTGCTCCGTCTAATGCCACTAATCAGGCTAATCTATGAACTCTGATCCGCCGCAGCTGTTCGTGCTCGCCGCCTTTGCCGCCTCCCAGGCTATCGCCCATCCCGG **************************************************.(((..(((((((.(((((......((((..(((((.....))))).....))))(((((...............)))))((((((((....)))))))).........(((........)))......)))))..)))))))...))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
M023 head |
SRR553488 RT_0-2 hours eggs |
M024 male body |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................TGCTCCGTCTAATGCCAC...................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................CAGCTGTTCGAGCTCGCC................................... | 18 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................TTTATCTCCCTCAGA........................................................................................................................................................................... | 15 | 1 | 7 | 0.29 | 2 | 0 | 0 | 2 | 0 | 0 |
| ...............................................................................................TTAAAGGCTGCGACAACC......................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................GTTTGTTGGTGGAATCGGTTT........................................................................................................................................................................................ | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 |
|
TAATTCAGATATAATGTTGGGTAACTTCGAGTATACTCAGTGGATCATACAACAACCCTAGCCAAAATAGAGTGAGTCTTATTCATTACATACGAAATTTCAGACGCTGTTGATTGGTTTAAGTGACAGCTCACCGTCGGTGGACGACGAGGCAGATTACGGTGATTAGTCCGATTAGATACTTGAGACTAGGCGGCGTCGACAAGCACGAGCGGCGGAAACGGCGGAGGGTCCGATAGCGGGTAGGGCC
**************************************************.(((..(((((((.(((((......((((..(((((.....))))).....))))(((((...............)))))((((((((....)))))))).........(((........)))......)))))..)))))))...))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................TCATTACATACGAAATTTCAGACGCT.............................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................AAACGGCGGAGGGGCC................ | 16 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................CAGATTACGTTGATT................................................................................... | 15 | 1 | 13 | 0.23 | 3 | 0 | 0 | 0 | 3 | 0 |
| ..........................................................................................TACGAAGTTTCAGAGGGTG............................................................................................................................................. | 19 | 3 | 20 | 0.20 | 4 | 0 | 0 | 4 | 0 | 0 |
| ...................................................................................................................................................................................................................................AGGGTCCGCTAGTGGGTA..... | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................TCAACGTCGGTGGAC......................................................................................................... | 15 | 1 | 20 | 0.15 | 3 | 0 | 1 | 2 | 0 | 0 |
| ..................................................................................................................................TCAACGTCGGTGGACCA....................................................................................................... | 17 | 2 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 |
| ............................................................................................................................................................................................................AGCAGGAGCGGCGGCAAGGGC......................... | 21 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................TCCGAAAGCGGGTAACGCC | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................TTTAAGTGTCAGGTCAC.................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................CCAGATTACGTTGATT................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................TCGATGGATGACGAGGTAG............................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................................................................................AGGGTCCCCCAGCGGGTAG.... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
Generated: 04/23/2015 at 10:47 PM