ID:dsi_11131 |
Coordinate:x:1613101-1613355 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
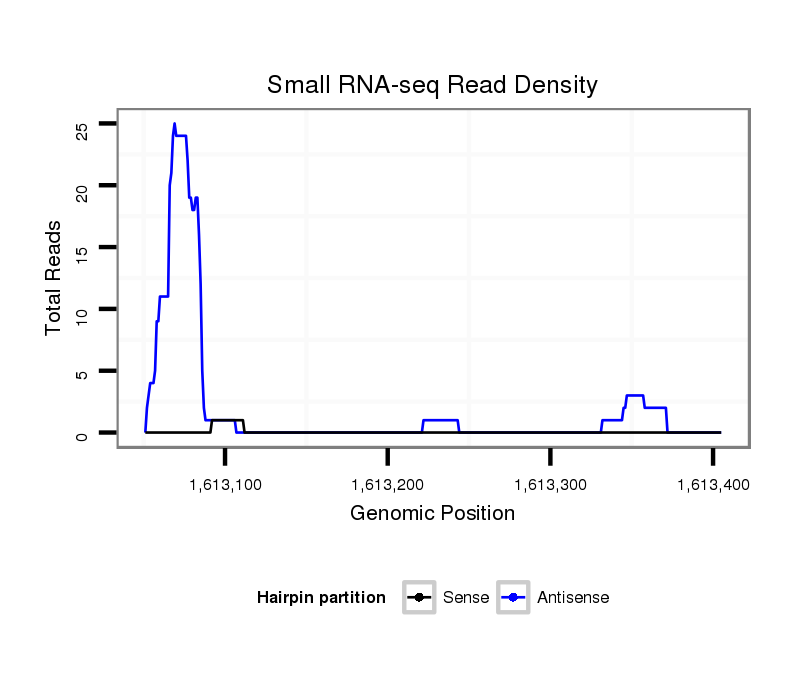
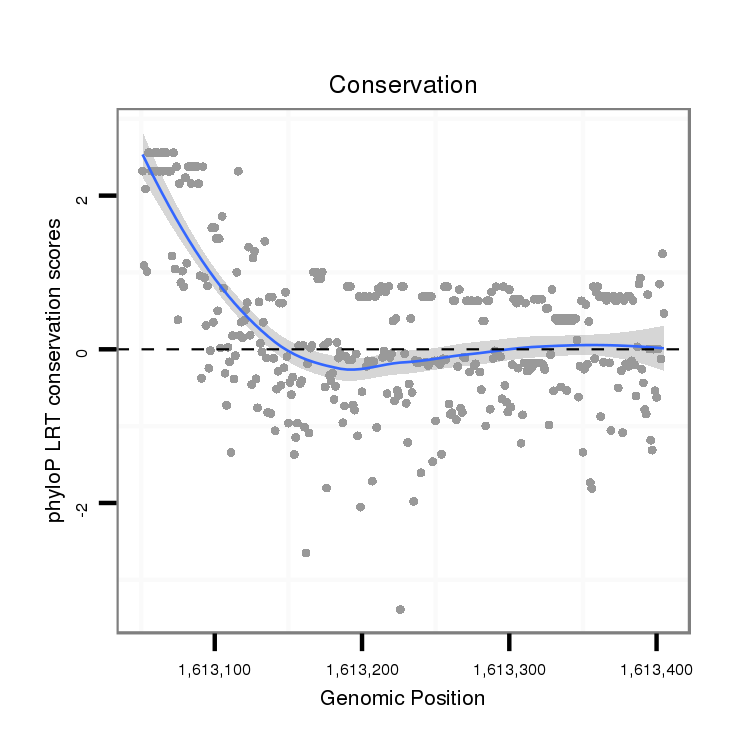
Antisense to five_prime_UTR [x_1612925_1613119_-]; Antisense to exon [x_1612922_1613119_-]; Antisense to three_prime_UTR [x_1613080_1613639_-]; Antisense to exon [x_1613080_1613955_-]
No Repeatable elements found
|
CCCGTAGCTGACGTGTTCGGTTCTGCGACAAATGTTTACCGTGGCTTTGTGTGAGTGCGGGTAGTGTGACCAGCGCAGTGTTAATGCGCCTTGGGCCAAAAGGGGTAAGCTTGTGGATACCCTGCCATTTTTTTTTGCTACTCTATTTGGAAAATTTTTATGTGTCTAGCCTTAGCAAGTTCACGACACCATCACTTCTTACCAGTGTTGAATTGAACATTTCAAGATTGTGCAAAGCATTTATTTAATAGGTTGTACCAGGCATATATTAACATATATAAACAAACATACTCTTTTCACTGTAGGAAGTAATGAGTCCAAGCATCGCCTTTACCAACACCGGCCGACACCGGCT
**************************************************(((((...(((.((((((.((((((((.......)))))..))).(((((.((((((.((..((((........))))........)))))))).)))))....((..((((((((((((((......(((((((((((...............))))))...)))))......((((.((((...))))..))))...))))).....)))))))))..))..............)))))))))))))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
M023 head |
M053 female body |
SRR902008 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR902009 testis |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................GTTCATGCGCCTTGGGCCG................................................................................................................................................................................................................................................................. | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................TGGCTTTGTGTGAGTGCGGG...................................................................................................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........TGTCGTGTTCGGTTGTCCGACA..................................................................................................................................................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................TGTGATTGCGGGTAG................................................................................................................................................................................................................................................................................................... | 15 | 1 | 7 | 0.71 | 5 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................TGTGATTGCGGGTAGG.................................................................................................................................................................................................................................................................................................. | 16 | 2 | 20 | 0.40 | 8 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................CAGTCTTTTCACTGTAGG................................................. | 18 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGTGAGTGGTGGTAGTGTG............................................................................................................................................................................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................TTTTGCGACTGTATTTGCAAAA......................................................................................................................................................................................................... | 22 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................ATAGATTCTTCCAGGCATAT........................................................................................ | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................CGACAAATGTTTACCGAGCT...................................................................................................................................................................................................................................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................GTGCGGGTAGTTTGACT............................................................................................................................................................................................................................................................................................ | 17 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................CTGTGATTGCGGGTAG................................................................................................................................................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................GTAGTGTGTCCAGCGCGA..................................................................................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................GCTATTTTTTTTTGCT........................................................................................................................................................................................................................ | 16 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ################################################################################################################################################################################################################################################################################################################################################################### GGGCATCGACTGCACAAGCCAAGACGCTGTTTACAAATGGCACCGAAACACACTCACGCCCATCACACTGGTCGCGTCACAATTACGCGGAACCCGGTTTTCCCCATTCGAACACCTATGGGACGGTAAAAAAAAACGATGAGATAAACCTTTTAAAAATACACAGATCGGAATCGTTCAAGTGCTGTGGTAGTGAAGAATGGTCACAACTTAACTTGTAAAGTTCTAACACGTTTCGTAAATAAATTATCCAACATGGTCCGTATATAATTGTATATATTTGTTTGTATGAGAAAAGTGACATCCTTCATTACTCAGGTTCGTAGCGGAAATGGTTGTGGCCGGCTGTGGCCGA **************************************************(((((...(((.((((((.((((((((.......)))))..))).(((((.((((((.((..((((........))))........)))))))).)))))....((..((((((((((((((......(((((((((((...............))))))...)))))......((((.((((...))))..))))...))))).....)))))))))..))..............)))))))))))))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M025 embryo |
SRR618934 dsim w501 ovaries |
M024 male body |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
O002 Head |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
SRR902008 ovaries |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............AAGCCAAGACGCTGTTTACA................................................................................................................................................................................................................................................................................................................................ | 20 | 0 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............AAGCCAAGACGCTGTTTAC................................................................................................................................................................................................................................................................................................................................. | 19 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......GACTGCACAAGCCAAGACGCTGTTTA.................................................................................................................................................................................................................................................................................................................................. | 26 | 0 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................GCCAAGACGCTGTTTACAA............................................................................................................................................................................................................................................................................................................................... | 19 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......GACTGCACAAGCCAAGACGCTGTC.................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .GGCATCGACTGCACAAGCCAAGACGC........................................................................................................................................................................................................................................................................................................................................ | 26 | 0 | 1 | 2.00 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................TACAAATGGCACCGAAACACACTCA........................................................................................................................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..GCATCGACTGCACAAGC................................................................................................................................................................................................................................................................................................................................................ | 17 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......CACTGCACAAGCCAAGAC.......................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................CCAAGACGCTGTTTACAAA.............................................................................................................................................................................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......GACTGCACAAGCCAAGACGCTG...................................................................................................................................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................AGTGACATCCTTCATTACTCAGGTT.................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........CTGCACAAGCCAAGACGCTGTTTACAA............................................................................................................................................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................AATCGTTCAAGTGCTGTGGTAG.................................................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................AGCCAAGACGCTGTTTACA................................................................................................................................................................................................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...CATCGACTGCACAAGCCAAGACG......................................................................................................................................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................CCCTGCATTAGTCAGGTTCGT............................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........CTGCACAAGCCAAGACG......................................................................................................................................................................................................................................................................................................................................... | 17 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................TGGGCACAACTTAACTTC......................................................................................................................................... | 18 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......CGACTGCACAAGCCAAGACGC........................................................................................................................................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................GCCAAGACGCTGTTTACA................................................................................................................................................................................................................................................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................TGTTTGTATGAGAAAAGTGACATCCT................................................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................AAAGTGACATCCTTCATTACTCAGGTT.................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................AGTGATGAGGCAGTGAAGA............................................................................................................................................................ | 19 | 3 | 7 | 0.43 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................TCAGGTTCGTGGCGG.......................... | 15 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................GCTTGTGGCCGGCTG....... | 15 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................................AAGTTCTAAAACGTTCCGGAA.................................................................................................................. | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................AAAAAAAAACGATGTTATGAAC.............................................................................................................................................................................................................. | 22 | 3 | 10 | 0.20 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| .................................................................................................................................................................................................................................................................................................................................CCTATCGGAAATGGTTGTGAC............. | 21 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................ATGATTGTGGCAGGCT........ | 16 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................GCTTGTGGCCGGCTGAG..... | 17 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................TGATTGTGGCAGGCTATG..... | 18 | 3 | 18 | 0.11 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .GGCATAGACTGTACAAGCG............................................................................................................................................................................................................................................................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................CTACATGGCTCAGGTTCGT............................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................TCAGGTTCGTAGCGT.......................... | 15 | 1 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................TCAGGTTCGTAGCGAGA........................ | 17 | 2 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................CTTCGGGACTCAGGTTCGT............................... | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................AGTCAGGTTCGTGGGGGAA........................ | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................CTGCATTAGTTAGGTTCGT............................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................TCAGGTTCGTACCGGC......................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................AAAAAAAACGACGAGATGA................................................................................................................................................................................................................ | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................TCATTCTTCAGGTTCGT............................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................CGTTGATTACTAAGGTTCG................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | x:1613051-1613405 + | dsi_11131 | C-----------------CCGTAGCTGACGTGTTCG------GTTCTG-CGAC-AAATGTTTACCGTGGCTTTGTGTGA------------------------GTGCG--GG---TAG------TG------------------------TGAC-CAGCGCA--------------------------------------GTGTTAATGCGCCTTGGGCCAA----AAGGGGTAAGCTTG-TGGATACCCTGC-CATTTTTTT---------------TTGCT-ACTCTATTTGGAAAATTTTTATGT--GTCT-AGCCTTAGCAAGTTCACGACACCA-TCACTTCTTACCAGTGTT-----------G-----------------------------------------------------------------------------------------------A---------------------ATTGAACATTTCAAGATTGTGCAAAGCATTTATTTAATAGGTTGTACCAGGCATATATTAACATATATAAACAAACATACTCTTT---------------TCACTGTAGGAAGTAAT-GAGTC--------------CAAGCATCGCCTTTACCAACACCG------------------------------------------------------------------------------------------GCCGACACCGGC--T |
| droSec2 | scaffold_10:1556252-1556605 + | C-----------------CCGTAGCTGACGTGTTCG------GTTCTG-CGAC-AAATGTTTACCGTGGCTTTGTGTGA------------------------GTGCG--GG---TAG------TG------------------------TGAC-CAGCGCA--------------------------------------GTGTTAATGCGCCTTGGGCCAA----AAGGGGTAAGCTTG-TGGATACCCTGC-CATTTTTT----------------TTGCT-ACTCTATTTGGAAAATTTTTATGT--GTCT-AGCCTTAGAAAGTTCACGACACCA-TCACTTCTTACCAGTGTA-----------G-----------------------------------------------------------------------------------------------A---------------------ATTGAACATTTCAAGATTGTGCAAAGCATTTATTTAATAGGTTGTACTAGGCATATATTAACATATATAAACAAACATACTCTTT---------------TCACTGTAGGAAGTAAT-GAGTC--------------CAAGCATCGCCTTTACCAACACCG------------------------------------------------------------------------------------------GCCGACACCGGC--T | |
| dm3 | chrX:1755757-1756135 + | C-----------------CCGTAGCTGACGTGTTCG------GTTCTG-CGAC-AAATGTTTACCGTGGCTTTGTGTGA------------------------GTTCG--GG---TAG------TG------------------------TGAC-CA-----GCGCCACTGC-------------------GGAAACACAGTGTTAATACGCCTTGGGCCAA-GGTAGGAGGTAGGCCCG-TGGATACCCTGC-CATTTTTTT------TTTTTTTTTTTGCT-ACTCCATCTAGAAAATTGTT--GT--GTCT-AGCCTTAGCAAGTACACCGCACTACTCACTTTTCACCAGTGTA-----------G-----------------------------------------------------------------------------------------------A---------------------ATTTAACATTTCAAGATTGTGCAAAGCAATTATTTAATAGGTTGTATTAGGCATATATTAACATACATAAACAGACATACTCTT----------------TCACTGT---AAGTAAT-GAGTC--------------CAAGCATCGCCTTTACCAACACCG---------------------------------------------------------------------------------------CCGGCCGACACCGGC--T | |
| droEre2 | scaffold_4644:1735008-1735387 + | C-----------------GCGTAGCTGACGTGTTCG------GTTCTG-CGACAAAATGTTTACGGTGGCCTTGTGTGA------------------------GTGCG--GG---TAG------TG------------------------TGAC-CA-----AAGCTACTGC-------------------GGAAACGCAGCGTTAATGCGCCGCTGACCAA----AGAGAATAAGCCCGTGGGATACCCTGG-AATTTTTT----------------GTATT-ACTGTATCTCGAAAATAATTATGT--GTCT-AGTCCTTTTAAGACTATTACACCATTCAGTTCTCATCAGTGTAAACC--CTTTAA-----------------------------------------------------------------------------------------------A---------------------ATTGAAAATTTCAAGATTGTGCAAAACATTTATTTCGTAAGTTGTACTAGGCATATATTAACT--CTTGAACAAACAAACTCTT----------------TCACTGTATAAAGTAAT-GAGTC--------------AAAGCTTTACCTTTACCAACACCG---------------------------------------------------------------------------------------ACAGCCGGCACCGGC--T | |
| droYak3 | X:1600772-1601149 + | C-----------------GCGTAGCTGACGTGTTCG------GTTCTG-CGCC-AAATGTTTACGGTGGCTTTGTGTGA------------------------GTGCG--GG---TAG------TG------------------------TGAC-CA-----AAGCCACTCC-------------------GGAAACGCAGCGTTAATGCGCCTCTGACCAA----AAAGATTAGGCC-GTGGGATACCCTGG-CATTATAT----------------TTATT-ACTGTGTCTGAAAAATTACTGTGT--GCCT-AGCCCTTTGAAAACTATTACACTATTCACTTGTCATCTGTGTAAATC--CTGTAG-----------------------------------------------------------------------------------------------G---------------------ATAGATGATTTCAAGATTGCGCCAAACATTTATCTCGCAAGTTGTACTAGGCATATGTTAACT--CTTAAACAAACATACTCTT----------------TTAATGAAACAAGTAGT-GAGTT--------------CAAGCTTTACATTTACCAACACCG---------------------------------------------------------------------------------------CCAGCCGGCACCGGC--T | |
| droEug1 | scf7180000409182:305292-305369 + | C-----------------GCGTAGCTGACGTGTTCG------GTTCTG-CGACAAAATGTTTACGGTGGCTCTGTGTGA------------------------GTGTG--AA--CTGA------TG------------------------TGAC-CA-----GAGC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBia1 | scf7180000301760:4359284-4359363 + | C-----------------GCGTAGCTGACGTGTTCG------GTTCTG-CGACAAAATGTTTACGCTGTGTGTGTGCGT------------------------GTGCG--TTGTGTGG------TG------------------------TGAC-CA-----GAGC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000415704:1361078-1361400 + | C-----------------GCGTAGCTGACGTGTTCG------GTTCTG-CGACAAAATGTTTACGCTGTGCGTGTGTG--------------------------TTG----A---AATAATCGATGTTGCTATCGGTGATATCGATGGTTGGGC-CC-----CGGCCACCGCCATCGATATTTTTTAGACTACTCTTACGCAGGAAA-----------------TGTGAGTGACGACTG-CGGGATACCCTGT-TTCGCGTTT---------------------------------------------------------------------------------------CGGGTATA-----------T----------------------------------------------AATTATACAT----------------------ATTTGT-ATACGTTTAAAATTTGCTCTAT-CATGTATGTATAGATCGATTAAAAGTTT---TAAACCTTTTTTTTATAA--------------------------TT-----------------TAATAGCAATAACTTACAGGGT---ACGT---------------------------------------GTACACTC------------------------------------------------------------------------------------------CCCCTTTTTGGC--A | |
| droEle1 | scf7180000491001:1258467-1258616 + | C-----------------GCGTAGCTGACGTGTTCG------GTTCTG-CGACAAAATGTTTACGCTGGCTGTGTGTGA------------------------GTGCG--GG--CCAG------TG------------------------TGAC-CA-----GAGTCGCCAC-------------------GCAAA-GCACCGTAAAATCTCTGTT---GATAGGTGGCGCAACGGCTTT-TGAATACCCTAC-TACTTACTT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000779506:1344993-1345419 - | C-----------------GCGTAGCTGACGTGTTCG------GTTCTG-CGACTAAATGTTTACGCTGGCTGTGTGTGA------------------------GTGCG--GA--CCGG------TG------------------------TGAC-CA-----GAGTCACAGC-------------------GCAAATGCACCGTAAAATCCCT---GGCGATGGGGTGCACACCGACTAG-ACGATACCCTGC-AACATACAAAGCTTTTTTTTTTTTTAACC-AATCCCT--------------TGTGAATCATAAC-C----AAGAT--------------------------------------------ATACTAATTTCTGACTACGAAAAGAAAATCAAGTCGGCTTACTAGTTATACAGAGACCATTATGAATTTATGCTCATTTATCCTACGCATAACTTTTCCTATGAAC------------AACAA--------------------------TACAGATTGGAGCTGTATGTTATTTATTTATTT-----------------AAATAGCCTTTACTTC-----------------------------------------------------TTCAATCAAAGGT---------------------------CAAATTTTGTATATGTCACCCTATACTACATTAAATTTATG-------------TTTTCCAGCGACGAC--T | |
| droFic1 | scf7180000454072:2867794-2868184 + | C-----------------GCGTAGCTGACGTGTTCG------GTTCTA-CGACAAAATGTTTACGGTGGCTGTGTGTGA------------------------GTGCG--GG--ACAG------TG------------------------TGAC-CA-----AAGCCACTGT-------------------GCATATTCACCGCAAA-----------------GCAGAGTATCGGTAA-CGGGATACCCTGCACATTTTTT----------------TTCTTAACTCTCCAT--AACTTTTTTTTGTAGGTCCCAGC-T----AAGTTC--CATTTTATTTACTTCATTCCTTTATCAGACATTTATTGTATATTTTA-TTTTTCAACAGGTAAAGAAAAACA------------------------------------------------------------------------------------------------------------TCATTCAGCTTTC-----------------------------------------------------------------------AAAGTATTTAAGTCAGATCTGAAACACTAAAGCATTCCCTGTTCCGACAACGAAGGGCTACTATTAATGTACCTACCAAACTCTACAAATTTCGT-----------------------------TGTTTTTCAACACCCAGAG-TTACACCGGC--T | |
| droKik1 | scf7180000302469:1171859-1171920 + | C-----------------GCGTAGCTGACGTGTTCG------GTTCTGGC-----GATGTTTACGGTGC----------------------------------GCGGT--GG--CCAG------TG------------------------TGGC-CA-----AC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13117:1056326-1056424 + | CAATCCTAAACCGAATCCG-CTAGCTGACGTGTTCG------GTTCTG-CGAC-AAATGTTTACGCTGGCTGTATGTGT------------------------GTTGC--GT---CTG------TG------------------------TGTCACA------GGCCA--------------------------------GTGTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBip1 | scf7180000396425:1058871-1058969 - | CAATCCTAAACCGAATCCG-CTAGCTGACGTGTTCG------GTTCTT-CGAC-AAATGTTTACGCTGGCTGTGTGTGT------------------------GTTGC--GT---CTG------TG------------------------TGACACAGACCAGAG--------------------------------------TT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| dp5 | XL_group1e:3768086-3768138 + | C-----------------GCGTAGCTGACGTGTTCG------GGTA---------------------------------------------------------GTTTGGTTA--CCAA------TG------------------------TGAC-CG-----TAGACT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAT | |
| droPer2 | scaffold_17:701567-701619 + | C-----------------GCGTAGCTGACGTGTTCG------GGTA---------------------------------------------------------GTTTGGTTA--CCAA------TG------------------------TGAC-CG-----TAGACT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAT | |
| droVir3 | scaffold_12970:1160686-1160776 - | C-----------------GCGTAGCTGACGTGTTCG------GTTCTG-CAACAAAATGTTTACGATTGCTT-------GCGTATGTGTGTAAGTGATGACAG---------------------CG------------------------CA---CAGCGCA--------------------------------------CTG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAAT | |
| droMoj3 | scaffold_6308:1236051-1236136 + | C-----------------GCGTAGCTGACGTGTTCG------GTTCTG-CAACTAAATGTTTACGATTGCGT-------GCGTATGTGTGCTAGAGATGGCAG---------------------CG------------------------CGAA-TAGCA----------------------------------------CAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15081:1760564-1760658 - | C-----------------GCGTAGCTGACGTGTTCGATATTGGTTCTG-CAAAGAAATGTTTACGATTGCGT-------GCGTATGTGTGTAAGTGA---------------------------TG------------------------ACAG-TAGCGCA-----A--------------------------------GTTTTGATG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/19/2015 at 05:49 AM