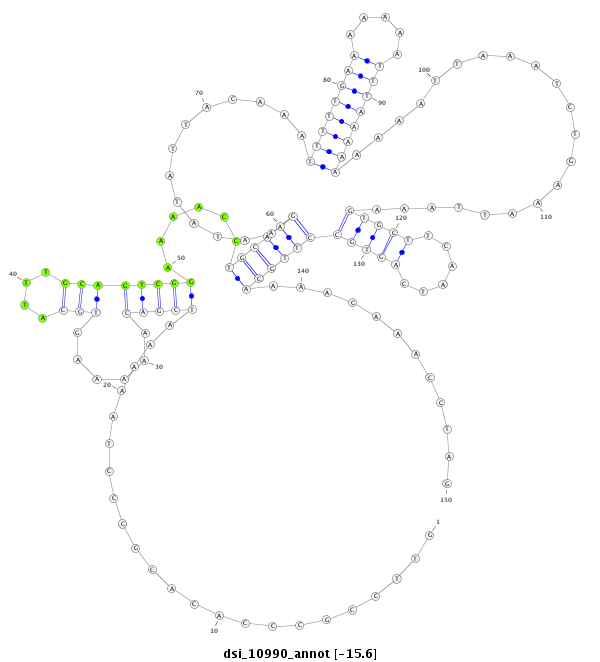
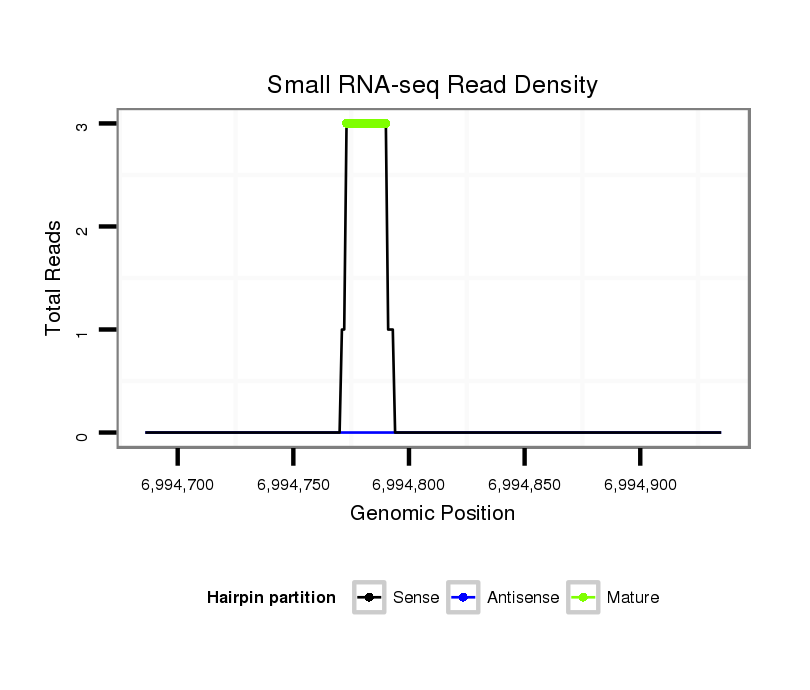
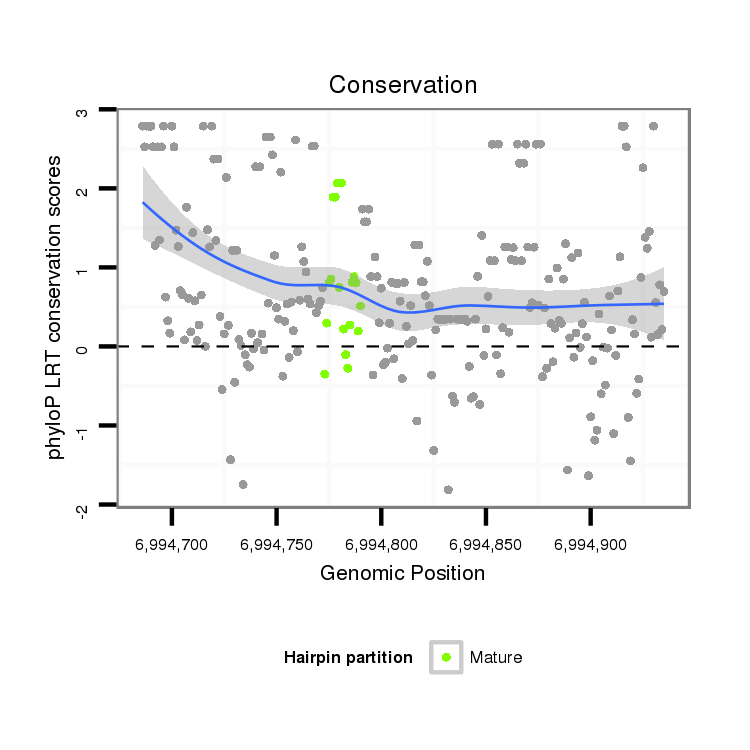
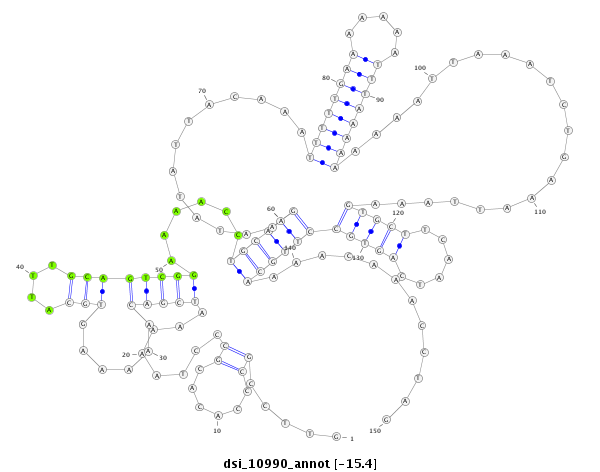
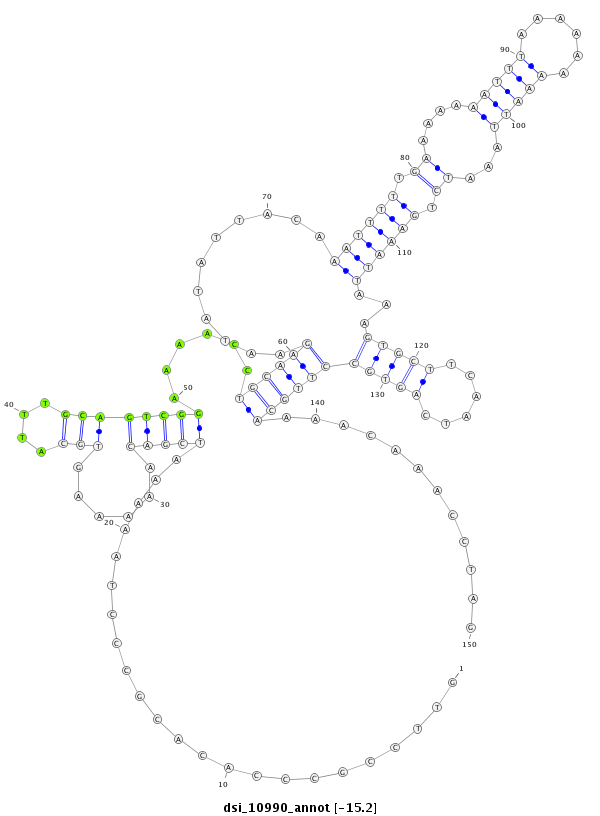
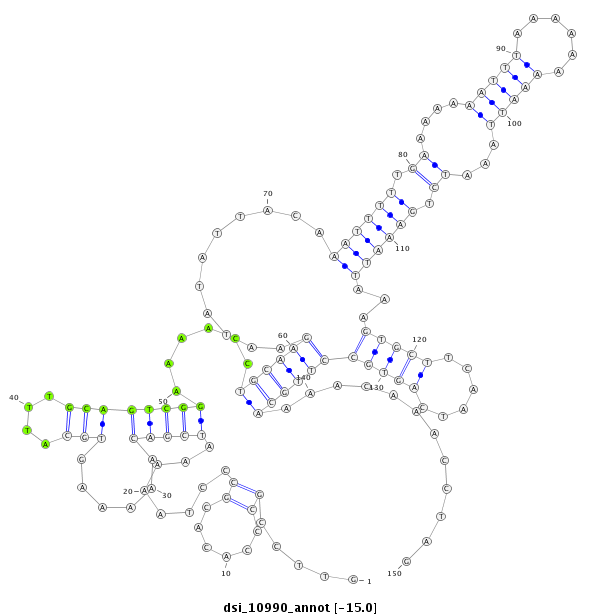
| droSim2 |
3r:6994686-6994935 + |
dsi_10990 |
TCTAA-CACAC--ACT----CACACA----CCCAAGAAA--CA--------CA----------------CA--------------------------------------------------------CACAC--------------ACA------------CATACAGAG---TTCCGCC-CACACGCCCTA---------------AA--------------------------------------------------------------------------------------------------------------------------------------------AATC--GACAAAAAGTGC-----ATTTGCAGTCG--------------------------------------------GA-A--AACCTGCAAGAATA-----------------------------------------------------------------------------------------TATT--A--------CAA----ATTTTTGAAAAAAATTTAAAAAAAAATTAAATCTGAAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTAAAGTGC-TTCA------AT--CAGTGCCTTGCAAA----AC------AAACCTAGAAT-------------------------------AAAATACCCCC--AGATA------------AAATATCCAGATCATTACT-GATCAGTGAA--GA---------------- |
| droSec2 |
scaffold_5:5622501-5622750 + |
|
TCTAA-CACAC--ACT----CACACA----CCCAAGAAA--CA--------CA----------------CA--------------------------------------------------------CACAC--------------ACA------------CATACAGAG---TTCCGCC-CACACGCCCTA---------------AA--------------------------------------------------------------------------------------------------------------------------------------------AATC--GACAAAAAGTGC-----ATTTGCAGTCG--------------------------------------------GA-A--AACCTGCAAGAATA-----------------------------------------------------------------------------------------TATT--A--------AAA----ATTTTTGATAAAAATT-AAAAAAAAATTAAATCTGAAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTAAAGTGC-TTCA------AT--CAGTGCCTTGCAAA----AC------AAACCTAAAAT-------------------------------AAAATACCCCC-CAGATA------------AAATATCTAGATCATTACT-GATCAGTGAA--GA---------------- |
| dm3 |
chr3R:14272615-14272868 - |
|
TCTAA-CACAC--ACT----CACACA----TCCAAGAAA--CA--------CA----------------CA--------------------------------------------------------CACAC--------------ACA----------CATATACAGAG---TTCCGCC-CACACGCCCTA---------------AAA-------------------------------------------------------------------------------------------------------------------------------------------AATC--GACAAAAAGTGC-----ATTTGCAGTCG--------------------------------------------AA-A--TACCTGCAAGAATA-----------------------------------------------------------------------------------------TATT--A--------AAA----ATTTTTGATAAAAATTAAAAAAATAAATAAATCTGAAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTAAAGTGC-TTCA------AT--CAGTGCCTTGCAAA----AT------AAACCTAGAAT-------------------------------AAAATACCCCC-CCCCCC------------CCTTATACAGATCGCTACT-GATCAGTGAA--GA---------------- |
| droEre2 |
scaffold_4770:7365925-7366134 + |
|
TCTAA-CACAC--ACT----CACACA----CCCAAGAAA--CA--------CA----------------CA--------------------------------------------------------CACAT--------------ACA------------------GAG---CTCCGCC-CACACGCCCTA---------------ATA-------------------------------------------------------------------------------------------------------------------------------------------AATC--GAGAAAAAGTGC-----ATTTGCAGTCG--------------------------------------------GA-A--AACCTGCAAGAATA-----------------------------------------------------------------------------------------TATT--A--------CAA----ATTTCTGATAAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAA--------AAGTGC-TTCA------AT--CAGTGCCTTGAAAA----AT------AAACCTAGA-T-------------------------------TAAATACCCCC-CAC---------------------CCAGATCGTTACT-GATCAGTGAA--GA---------------- |
| droYak3 |
3R:2905614-2905853 - |
|
TCTAA-CACAC--ACT----AACACA----CCCAAGAAA--CA--------CA----------------CA--------------------------------------------------------CGCAC--------------ACA------------------GAG---CTCCGCC-CACACGCCCTA---------------AAA-------------------------------------------------------------------------------------------------------------------------------------------AATC--GACAAAAAGTGC-----ATTTGCAGTCG--------------------------------------------GA-A--AACCTGCAAGAATA-----------------------------------------------------------------------------------------TATT--A--------CAA----ATTTTTGATAAAA-----AAAAAGAGTTAAATCTATAATTTAAAAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTAAAGTGC-TTCA------AT--CAGTGCCTTGAAAA----AT------AAACCTAGAAT-------------------------------AAAATACCCCT-TAC---------------------CCAGATCATTACT-GATCAATGAA--GA---------------- |
| droEug1 |
scf7180000409802:1767721-1767975 + |
|
TCTAA-CACAC--ATT----CACACA----CCCAAGAAAACTA--------CA----------------CA--------------------------------------------------------CACAT--------------ACA------------AAAACACAG---TTCCGCC-TACACGCCCTA---------------AAA-------------------------AAATAG------C---------------AACCA------------------------------------------------------------------------------AT-AATTGCAAC-AAAAGTGC-----TTTTGCAGTCG--------------------------------------------GA-A--AAACTGCAATAAAA-----------------------------------------------------------------------------------------AATT--A--------AAA----ATTTTTAATA------------------------------------------AGAAAAT----------------------------------------TT------------------------------------------------------------------------------------------------------TT---------------------------------------AAAA-------ATTAAGTGC-TAAA------AT--CAGTGCCTTGAAAA----AT-AT-ACATACTTAATAT-------------------------------AAATTACCCCT--AA---------------------ACAGATCAAAATT-GATCAGTGAATTGAA-------------AG |
| droBia1 |
scf7180000299102:1684448-1684703 + |
|
TCTAA-CACAC--ACT----CACACA----CCCAAGAAA--CA--------CA----------------CA--------------------------------------------------------CACAC--------------ACA------------------TAG---TTCCGCC-CACACGCCCAC---------------T--------------------CCAAGAAAGAGAG-------------------CCCAAAAA---------------------------------------------------------------------------------AATC----------GTGC-----ATTTGCAGTCG--------------------------------------------GGAA--AAACTGCAAAAAAA-------A---------------------------------------------------------------------------------TTATTTA--------AAAATATATTTTTAATA---ATC-----------------A------------------AAAAATTA--AA------------------------------G---------------------------------------------------------------------------------------------------------------T---------------------------------------------------TTGTGCCTAAA------AT--CAGTGCCTTGAAAATAAAAT-AC---ATACTTCAAAT-------------------------------TAAACACCCCT-C-C--A----AC------CCTCA-AAAGATCCGTATC-GATCAGTGAA--GAA--------------- |
| droTak1 |
scf7180000415878:553475-553718 + |
|
TCTAA-CACAC--ACT----CACACA----CCCAAGAAA--CA--------CA----------------CA--------------------------------------------------------CACAC--------------ACA------------------TAG---TTCCGCC-CACACGCCCTC---------------AAAAATAT----ATGAAAAACC------------------C---------------AACAA------------------------------------------------------------------------------AT-AATTGCAAC-TAAAGTGC-----TTTTGCAGTCG--------------------------------------------GA-A--AAACTGCATAGAAA-------A---------------------------------------------------------------------------------TA----A--------AAA----AATATTA------A------------------------------------TTAAAAAATA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AATA--------AAGTGT-TAAA------AT--CAGTGCCTTGAAAA----AT-AT---ATACTTAAAAT-------------------------------TCAATACCCCC-TCC---------------------CCAGATCATTGAC-GATCAGTGAA--GAG--------------- |
| droEle1 |
scf7180000491008:1974113-1974356 + |
|
TCTAA-CACACACACT----CACACA----CCCAGGAAA--CA--------GT----------------CA--------------------------------------------------------CACAC--------------ACA------------------CAG---TTCCGCC-AACACGCCCAC---------------AAA---------------AACC------------------C---------------AACAG------------------------------------------------------------------------------AT-AGTTGCAGC-AAAAGTGC-----ATTGGCAATCA--------------------------------------------A-----AAATTGCAATAAAA-----------------------------------------------------AA-ATGTTTTGAAAT---------------------------------------------TT-------------------------------------CAAATCAAATTAGAAAATA--AA-----------------------------------T----------------------------------------------------------------------------------------------------------T--------------------------------------TAAAA--------AAGTGC-TAGA------AT--CAGTGCCTTGAAAA----A--------AACTTAAAAT-------------------------------CAATTACCCCT-CCC---------------------CCAGATCATTACC-GATCAGTGAA--GAA--------------- |
| droRho1 |
scf7180000777997:4681-4922 - |
|
TCTAA-CACAC--ACACACTCACACA----CCCAGGAAA--CA--------AA----------------AA--------------------------------------------------------CACAC--------------AC--------------------AG---TTCCGCC-CACACGCCCAC---------------AAAT---------------ACC------------------C---------------AACAG------------------------------------------------------------------------------AT-AGTTGCAAC-AAAAGTGC-----ATTTGCAGTCA--------------------------------------------AAAA--AAACTGCAAAAAAA-----------------------------------------------------------------------------------------TTTTCCA--------AAAAT--AATTTTAAAAAA----------------------------------------------------------------------------------GT---T-------------------------------------------------------------------------------------------------------TT---------------------------------------AAAAAA--------AAGTGA-TTTA------AT--CAGTGCCTTGAAAA----AA--T---ATACTTAAAAT-------------------------------TAAATACCCCCTCCC---------------------CCAGATCATTATC-GATCAGTGAA--GTA--------------- |
| droFic1 |
scf7180000453800:750541-750789 - |
|
TCTAA-CACAC--ACT----CACACA----CCCAAGAAC--CA--------CA----------------CA--------------------------------------------------------CACAC--------------AC--------------------AG---TCCCGCC-CACA-GC------------------------------------AAACC------------------C---------------AACGG------------------------------------------------------------------------------AT-TATTGCAAC-AAAAGTGC-----ATTTGCAGTCG--------------------------------------------GT-T--AAACTGCAAAATC---------------------------------------------------AATACTTAGTTTTAGAAATTCAAA--------------------------TAAAAAA----ATATTTGAAAAAA---------------------------------------------------------------------------------GT---TT------------------------------------------------------------------------------------------------------TT---------------------------------------CAAAAT--------AAGTGC-TAAA------AT--CAGTGCCTTGAAAA----AT-AT---ATACTTAAATT------------------------------TAAAATACCCCT-C-C---------------------CCAGATCGTTACT-GATCAGTGAA--GA---------------- |
| droKik1 |
scf7180000302392:131058-131416 + |
|
TCTAAACTCAC--ACA----TACTCT--GAC-----ACA--CACTCACGCACACCCACGCCCACACACGCA--------------------------------------------------------CACAC--------------CCT------------T------GG---AT----------CGGG---------------------------------------C------------------C---------------AGTAA------------------------------------------------------------------------------AT-AATTGCAAC-AAAAGTGCGGCGAAATTGCAGTCG--------------------------------------------GAAA--AATCTGCAAGAAAA------------------------------------------------------------------------------------------ATC--A--------AAA----AATTGAAACA------------------------------------------ACAAAAT----------------------------------------TTTTCAACTAAAAAGTCTAGTTTGGCAAAATTACAGAAATTTTGAAATACAAATTTTGCCAAATAGGAATCATGAAATAAATAATAATTAATCATAAAAAATTTTTATTA-----------------------------------CAAAAA--------TAGTGC-TGAA------AT--CAGTGCCTTAAAAA----AG----------CCA---T-------------------------------TAAATAAA----T----ACATAATATTTTAACGCA-GCAGATCACCAAT-GATCAGTGCTGAGA---------------- |
| droAna3 |
scaffold_13340:9832373-9832594 + |
|
TCTAA-CACAC--ACC-A--CACACACAAACACCAGCAG--CA--------CA----------------CA--------------------------------------------------------CAC--------------------------------------AACACCATCGCAACATACGCGCCA---------------AAA------------------------AA-----------A---------------AGAAG------------------------------------------------------------------------------GA-AATTGTAAC-GAAAGTGCTAAACAACTGCAGTAA--------------------------------------------AAAAA-AGTCTGCAAAAAAAT-----AA---------------------------------------------------------------------------------ATACCCA--------CAA-----------------------------------------A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATTGTGT-TAAA------AG--TAGTGCCTTAAAAA----AT------AATTT-----------------------------------------TATCCCT-A-C--A------------ACATA-TCAGATCAATATTGGATAAGTGAA--GA---------------- |
| droBip1 |
scf7180000396691:2533654-2533868 + |
|
TCTAA-CACAC--ACT----CACACACAAACCCCAG-----CA--------CA----------------CA--------------------------------------------------------CACCA--------------C---------------------CA---CACCGCAACATACGCGCCA---------------AAA------------------------AA-----------------------------AAG------------------------------------------------------------------------------GA-AATTGTAAC-GAAAGTGCTACACCACTGCAGTAA--------------------------------------------AAAAAGAATCTGCAAAAAAT------AA---------------------------------------------------------------------------------TTACCCA--------AAA-----------------------------------------A-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATTGTGC-TAAA------AC--TAGTGCCTTAAAAA----AT------AA--------T-------------------------------TGAATCCCCAC------A------------ACATA-TCAGATCAATATTGGATAAGTGAA--GA---------------- |
| dp5 |
2:11922230-11922598 + |
|
TCTAAGCACAC--ACT----CACACA--AAC-----TTA--CACGCGCGCACACACACACACACACGCACA------------A-----------------CACACCGTA----------------------CTCGGTGCA---------------------------------TACTCC-CACACGCACAC---------------AAATACGCGCGCGCGAAAAACC------------------C---------------AACCGCTTGGGAAAACACGGAAAAACAAAAACTTGGATACTTTTCAAACCAAAATATACACAACAAAAAAGTTGAAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAAAT-------------TT-------------------------------------CAAATTTCATAAGAAATTG--CA------------------------------G---------------------------------------------------------------------------------------------------------------TTCCCCCCCCTCCACCAAAAAAAAAAAAAACTGCTCGG-CAAAA--------AAGTGC-TAAAATCGAAAT--CTGTGCCTTAAAAT----A----------CCCAAAATATTTTGCCCATAAAAATACAAAATCAAATCA--------------------AAATTCTGT-------AGCGATCACTGTG-GATCGTTG-A--GA---------------- |
| droPer2 |
scaffold_0:8584309-8584671 - |
|
TCTAAGCACAC--ACT----CACACA--AAC-----TTA--CACGCGCACACACACACACACACA----CA------------A-----------------CACACCGTA----------------------CTCGGTGCA---------------------------------TACTCC-CACACGCACAC---------------AAATACGCGCGCGCGAAAAACC------------------C---------------AACCGCTGGGGAAAACACGGAAAAACAAAATCTTGGATACTTTTCAAACCAAAATATACACAACAAAAAAGTTGAAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAAAT-------------TT-------------------------------------AAATTTTCATAAGAAATTG--CA------------------------------G---------------------------------------------------------------------------------------------------------------TTCCCCCCCCTCCAGCAAAAAA--AAAAAACTGCTCGG-CGAAA--------AAGTGC-TAAAATCGAAAT--CTGTGCCTTAAAAT----A----------CCCAAAATATTTTGCCCATAAAAATACAAAATCAAATCA--------------------AAATTCTGT-------AGCGATCACTGTG-GATCGTTG-A--GA---------------- |
| droWil2 |
scf2_1100000004943:10759022-10759325 + |
|
TCTAA-C-CAC--ACT----TACATA----TACAT-ATA--TACA------TA----------------CAAATATATATATATATAT--------------------------ATATATATACGCACAC--AACAATACACAC--TCC------------------TTC---TCCCTCT-CACACGCACAC---------------GCA--------TGCT------------------------------GGGATT-ATT----------------------------------------------ACATCAAAACCCTTCTAAGAAAGAAGAAGAATAAATAAAATATTA--TACAACAAATCT-----TTTTGCAGTTTTTATGCAAAAAACATAAATCAAGTGCCGCCTACAAGTGCCTGAAAAAAT-AAT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTACA-AAAAA-------------------------------AAAAAATCATT-TAT---------------------ATAAATCTATGAA-AATCAGTGCT--AAATAATAATTAATTTAA |
| droVir3 |
scaffold_12855:4614645-4614966 + |
|
TCTAAGCACGCACACT----TACACT--AGC-----ACA--CACACACACACAC----------A----CAAACA--------ACTATACGCACGTATGCACACATCGTATAAAATATAAA------CACGC-ACATTACACACACACA--CACACACACACACACACAC---ATACGCA-CATAC---------------------------AT----GT-------------------------------GATGAT-TCCCAATAA------------------------------------------------------------------------------GA-TCTAGTAAC-AAAATA------------------------------------------------------------------------------A----A------------------------------AATAATCAATTAAAGAACAATAT-TTGTTCTAGAATTTCGCA--------------------------AAAA------------------------------------------------------------------GTGCAAAATATATGT--ATGCATATACACATACATATATAT----------------------------------------------------------------------------------------------------------T--------------------------------------GATAA--------AAGTGT-TTAA------ATTACAGTGCCTTACAAT----AAT-----AA--------------------------------------------------------------------------A-TGAAATCGGCTT----ACAATTAA--GG---------------- |
| droMoj3 |
scaffold_6540:8979046-8979370 + |
|
TCTAAGCACGCACAAT----TACACT--AGC-----ACA--CATACATGCATACATACGTACATATACACA--------------------------------------------------------CACAT--------------ACA------------TATATCGAG---T-----A-TACACGCACAC---------------ACAT---------------AGC-----CAACGCACACAGGCAACTGAAAATTTGCCAATAA------------------------------------------------------------------------------GA-TCTAGTAAC-AGAA------------------------------------------------------------------------------AAAAAA-------AAATAATAATAATAATTAAAATTCAAATAATAAATAAAAGAACAATAT-TTGTTCTAGAATTTCGCA--------------------------TAAA------------------------------------------------------------------GTGCAAATTATATGTCTATGCAT--------------ATAT----------------------------------------------------------------------------------------------------------T--------------------------------------GATAA--------AAGTGT-TTAA------ATTACAGTGCCTTACAAT----AAT-----AA--------T-------------------------------CGGCTACA----T----ACA-----------TATG-TGAAATCTGTTT----ACAATCAA--TG---------------- |
| droGri2 |
scaffold_14906:7133263-7133555 + |
|
TCTAAGCACGCACACT----TACACT--AGC------CC--CA--------CA----------------CA--------------------------------------------------------CACAT--------------ACATA----------C-T---ACA------CACA-TACACATACTCAAACGCATAATACACAGATACACGCGCGAT------------------------------AATGTT-GTCCAATCA------------------------------------------------------------------------------GA-TCTTGTAAC-AAAATC------------------------------------------------------------------------------A----AAAATAA---------------------------------TTGATGAACAATAA-TTGTTCTAGAATTTCGCAAAAAAAAAAAAAAAAAA----A--------ATT----TATATTA------A------------------------------------TTGAATAGTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTAAAATAATTAAATAT-TGAT------TG--CAATGTCTACCGAA----AT-TGGGTTTAATTAATAT-------------------------------AAATAA-------------T-----------TATA-TGCAATCAATTT----ACAGTTTA--AG---------------- |