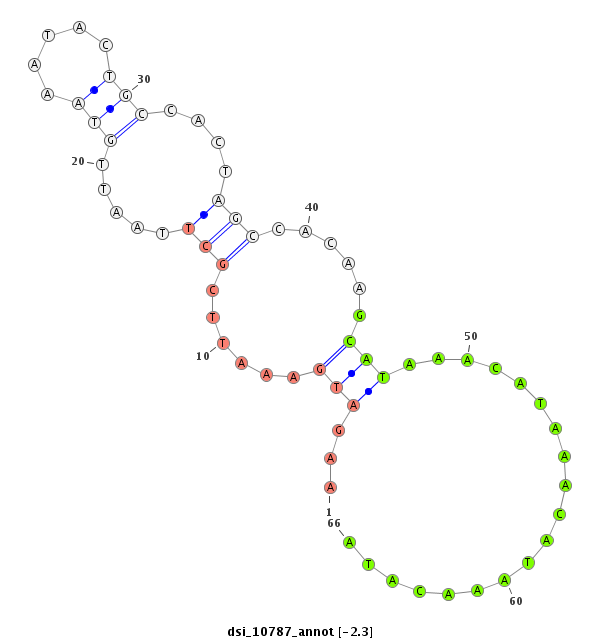
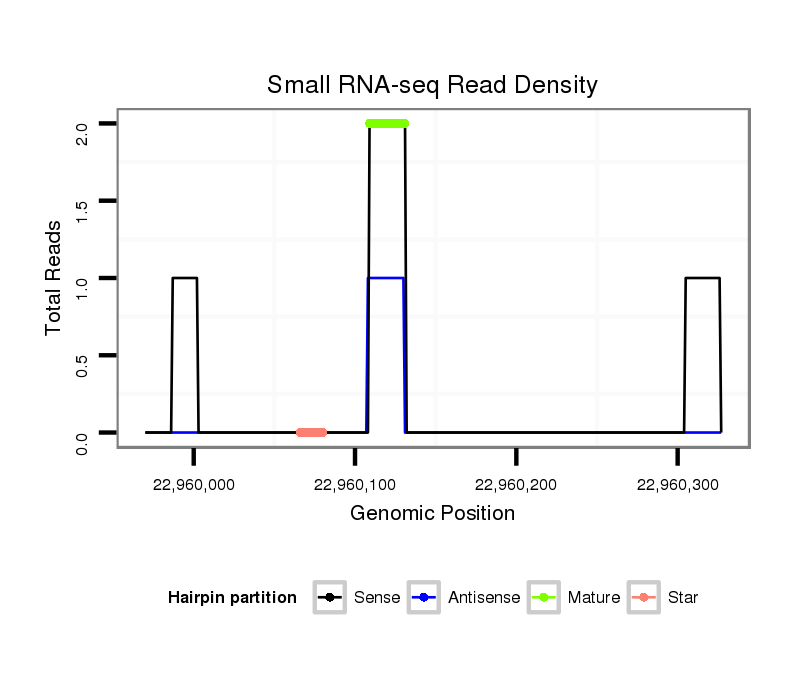
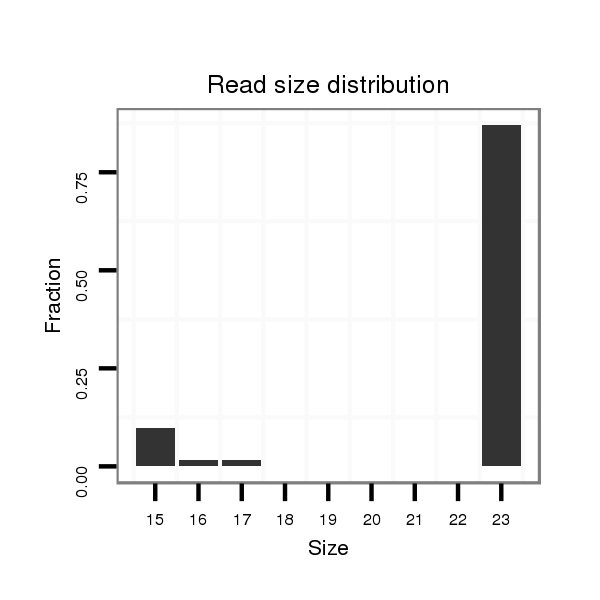
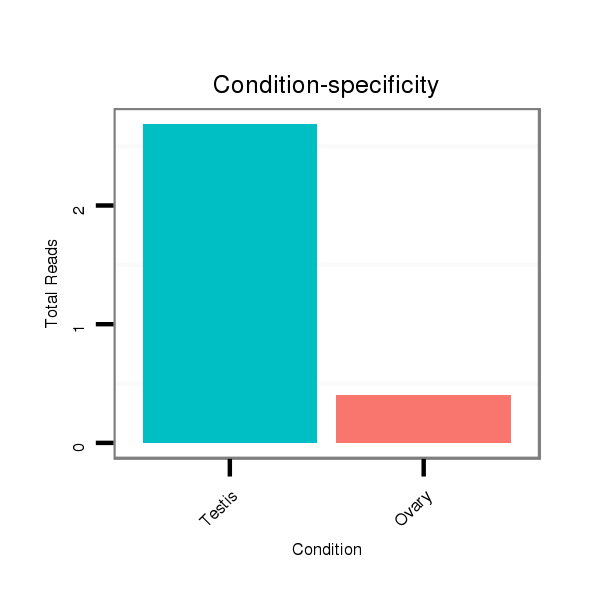
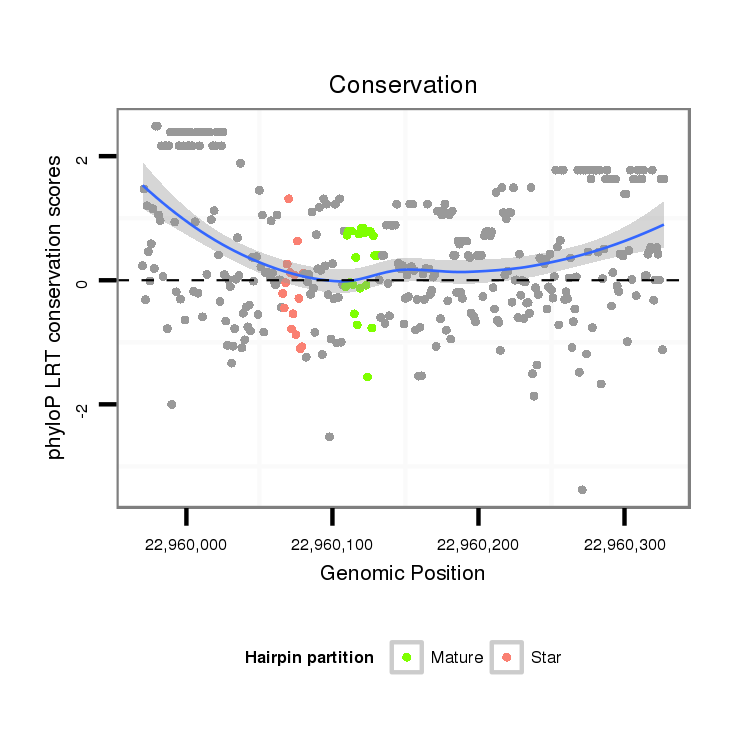
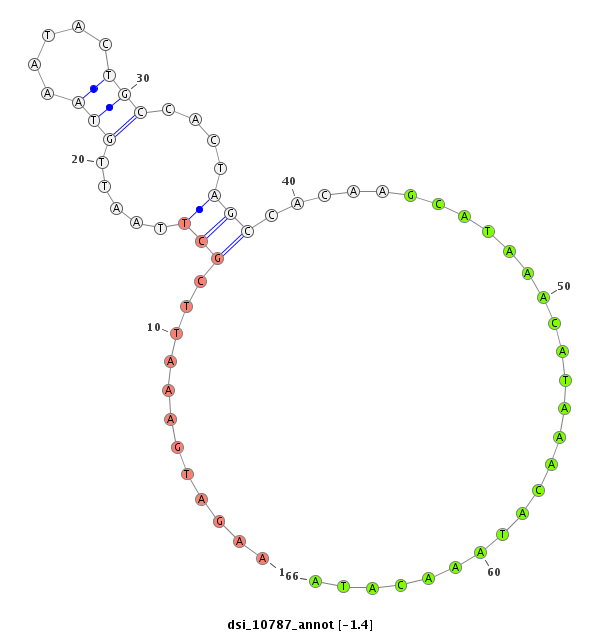
ID:dsi_10787 |
Coordinate:3r:22960020-22960277 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -1.4 |
 |
exon [3r_22960278_22960871_+]; CDS [3r_22960278_22960871_+]; exon [3r_22959509_22960019_+]; CDS [3r_22959509_22960019_+]; intron [3r_22960020_22960277_+]
No Repeatable elements found
| ##################################################------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CTGGGCGGCAAGCGGGCGCTCACCACATCCTGGAGCGTAGAGAAAAAGGGGTGGGTTTGTATACTAAACTATAGTTATTGTCTGTATGCACATTATAAGATGAAATTCGCTTAATTGTAAATACTGCCACTAGCCACAAGCATAAACATAAACATAAACATAATTTCTCCTTATGACTTCACATTTCTTCCTGGAAGTCATGAATGTTAACTTTTATGACATTTGCACGTATTTCTAACATCTCAATTGCATATAAAAAGTTTTTAATATCTTTACCCTAACAAACATTTAACATATATCTGTTGCAGAAAACTGAGCTCCTTGGGCAACGGCGAACCTTTCGATGCTTTAGACCCTG ************************************************************************************************...(((......(((.....(((.....)))....)))......)))...................**************************************************************************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
M053 female body |
M024 male body |
M023 head |
SRR902009 testis |
O001 Testis |
SRR553486 Makindu_3 day-old ovaries |
M025 embryo |
GSM343915 embryo |
SRR553485 Chicharo_3 day-old ovaries |
SRR902008 ovaries |
SRR618934 dsim w501 ovaries |
O002 Head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................................................................................................................................CTATCGATCCTTTAGTCCCT. | 20 | 3 | 2 | 26.00 | 52 | 6 | 5 | 15 | 14 | 8 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................CCTATCGATCCTTTAGTCCCT. | 21 | 3 | 2 | 16.00 | 32 | 13 | 9 | 4 | 0 | 2 | 0 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................CCTATCGATCCTTTAGACC... | 19 | 2 | 2 | 5.00 | 10 | 6 | 1 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................CCTATCGATCCTTTAGA..... | 17 | 2 | 14 | 3.14 | 44 | 20 | 13 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................TCCTGGAGCGTAGAGAAAAAGGGAAA................................................................................................................................................................................................................................................................................................................. | 26 | 3 | 1 | 3.00 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................TCGATCCTTTAGACC... | 15 | 1 | 7 | 2.71 | 19 | 17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................GCATAAACATAAACATAAACATA.................................................................................................................................................................................................... | 23 | 0 | 16 | 2.69 | 43 | 0 | 0 | 0 | 0 | 0 | 0 | 43 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................CTATCGATCCTTTAGTCCC.. | 19 | 3 | 5 | 2.40 | 12 | 4 | 0 | 1 | 2 | 3 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................CCTATCGATCCTTTAGACCT.. | 20 | 3 | 7 | 2.14 | 15 | 8 | 1 | 0 | 2 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................TATCGATCCTTTAGTCCCT. | 19 | 3 | 12 | 2.08 | 25 | 1 | 1 | 4 | 4 | 14 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................CCTATCGATCCTTTAGTCCC.. | 20 | 3 | 3 | 2.00 | 6 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................................................................................................................................................................................................CCTATCGATCCTTTAGACCTT. | 21 | 3 | 4 | 1.75 | 7 | 3 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................CCTATCGATCCTTTAGAC.... | 18 | 2 | 2 | 1.50 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................ACCTTTCGATGCTTTAGACCCT. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................TCGATGCTTTAGACCTT. | 17 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................GCTCACCACATCCTGG..................................................................................................................................................................................................................................................................................................................................... | 16 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................TGGAGCGTAGAGAAAAAGGGGTTT................................................................................................................................................................................................................................................................................................................ | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................GCTCATCACATCCTGGA.................................................................................................................................................................................................................................................................................................................................... | 17 | 1 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................CTTAGGACTTCACATT............................................................................................................................................................................. | 16 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................TCCTGGAGCGTAGAGAAAAAGGGTTT................................................................................................................................................................................................................................................................................................................. | 26 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................TCGATCCTTTAGTCCCT. | 17 | 2 | 7 | 0.86 | 6 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................TCGATCCTTTAGTCCCTC | 18 | 3 | 20 | 0.60 | 12 | 1 | 0 | 0 | 5 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................TATCGATCCTTTAGTCCC.. | 18 | 3 | 20 | 0.55 | 11 | 0 | 0 | 1 | 2 | 6 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................ACATAAACATAAACATAAACATAGTT................................................................................................................................................................................................. | 26 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................TTCGTGGCTTTAGACCC.. | 17 | 2 | 9 | 0.33 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................TAAGATGAAATTCGCTCACT................................................................................................................................................................................................................................................... | 20 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................CCTATCGATCCTTTAGTCC... | 19 | 3 | 14 | 0.29 | 4 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................................................................................................................................................................................................CTATCGATCCTTTAGAC.... | 17 | 2 | 11 | 0.27 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................ATCGATCCTTTAGACC... | 16 | 2 | 20 | 0.25 | 5 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................AAGATGAAATTCGCT....................................................................................................................................................................................................................................................... | 15 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................CCTTTCGATCCTTTAGAACTT. | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................CCTAACGATCCTTTAGACC... | 19 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................CTATCGATCCTTTAGCCCCT. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................CTATCGATCCTTTAGACC... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................CCTGGTGCGTAGAGA........................................................................................................................................................................................................................................................................................................................... | 15 | 1 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................CTATCGATCCTTTAGACCTT. | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......GCAAGAGGGCGATCACC.............................................................................................................................................................................................................................................................................................................................................. | 17 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CACGTTATAAGATGA............................................................................................................................................................................................................................................................... | 15 | 1 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................TATCGATCCTTTAGACC... | 17 | 2 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................GCGTAGAGCAAAAGG..................................................................................................................................................................................................................................................................................................................... | 15 | 1 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................TATCGATCCTTTAGAC.... | 16 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................TCGATCCTTTAGACCT.. | 16 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................CGATCCTTTAGTCCCT. | 16 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................GAGAAAAAGGGCTGAGTT............................................................................................................................................................................................................................................................................................................. | 18 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................GCGTAGAGCAAAAGGG.................................................................................................................................................................................................................................................................................................................... | 16 | 1 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................TCGATCCTTTAGACCTT. | 17 | 2 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................ATTATAATCTGAAATTCG......................................................................................................................................................................................................................................................... | 18 | 2 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................GTTCGTTGCTTTAGACC... | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................GCCTATCGATCCTTTAGAC.... | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................ACATAAACATAAACA...................................................................................................................................................................................................... | 15 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................CCTATCGATCCTTTAGAG.... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................AACATAAACATAAACA...................................................................................................................................................................................................... | 16 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................................................................................................CTTGGGCAGCTGCGAAC..................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................AACTGATTTGACATTTGCAC.................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................ACATAAACATAAACATA.................................................................................................................................................................................................... | 17 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
GACCCGCCGTTCGCCCGCGAGTGGTGTAGGACCTCGCATCTCTTTTTCCCCACCCAAACATATGATTTGATATCAATAACAGACATACGTGTAATATTCTACTTTAAGCGAATTAACATTTATGACGGTGATCGGTGTTCGTATTTGTATTTGTATTTGTATTAAAGAGGAATACTGAAGTGTAAAGAAGGACCTTCAGTACTTACAATTGAAAATACTGTAAACGTGCATAAAGATTGTAGAGTTAACGTATATTTTTCAAAAATTATAGAAATGGGATTGTTTGTAAATTGTATATAGACAACGTCTTTTGACTCGAGGAACCCGTTGCCGCTTGGAAAGCTACGAAATCTGGGAC
****************************************************************************************************************************************************************************************************...(((......(((.....(((.....)))....)))......)))...................************************************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR902008 ovaries |
SRR553485 Chicharo_3 day-old ovaries |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................AGTGTGACGAAGGAACTTCAGT.............................................................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................TGACTCGATGAACCCG............................... | 16 | 1 | 3 | 1.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................TCGTATTTGTATTTGTATTTGTA..................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................TGTATTTGTATTTGTA..................................................................................................................................................................................................... | 16 | 0 | 20 | 0.45 | 9 | 2 | 7 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................GTGAAGAAGGACCTTCA................................................................................................................................................................ | 17 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TTGTTTTAAACAGGAATA........................................................................................................................................................................................ | 18 | 2 | 16 | 0.25 | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................TTTGTATTTGTATTTGTA..................................................................................................................................................................................................... | 18 | 0 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................GACTCGATGAACCCG............................... | 15 | 1 | 12 | 0.17 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................TTGACTCGATGGACCCG............................... | 17 | 2 | 19 | 0.16 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................GTATTTGTATTTGTATTTGTA..................................................................................................................................................................................................... | 21 | 0 | 20 | 0.15 | 3 | 0 | 2 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................CGTATTTGTATTTGTATTTGTA..................................................................................................................................................................................................... | 22 | 0 | 17 | 0.12 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................TGTATTTGTATTTGTAT.................................................................................................................................................................................................... | 17 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TTGTATTACTGAGGAAT......................................................................................................................................................................................... | 17 | 2 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................CGTATTTGTATTTGTATTTGTAT.................................................................................................................................................................................................... | 23 | 0 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................ATTTGTATTTGTATTTGTAT.................................................................................................................................................................................................... | 20 | 0 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................GTATTTGTGTTTGTATTTGTA..................................................................................................................................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................ATTTGTATTTGTATTTGT...................................................................................................................................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................TGCTTTCAATTGAGAATAC............................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................AGTATTTGTATTTGTATTTGTA..................................................................................................................................................................................................... | 22 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................TAGATTGTATATAGA......................................................... | 15 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................GTATTTGTATTTGTATTTGT...................................................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................CGTATTTGTATTTGTA........................................................................................................................................................................................................... | 16 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................GGGAATGCTACGAAAT....... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................ATCTGTTGTTCCCCAC................................................................................................................................................................................................................................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3r:22959970-22960327 + | dsi_10787 | CTGGGCGGCAAGCGGGCGCTCACCACATCCTGGAGCGTAGAGAAAAAGGGGTGGGTTTGTATACTAAACTATAGTTATTGTCTGTATGCACATTAT-AA--------------------------GATGAAATTCGCTTAATTGTA----AATACTGCCACTA-GCCA-CAAGCATAAACATAAACATAAACATAATTTCTCCTTATGACTTCACATTTCTTCCTG----GAAGTCA------------------TG-AATGTTAACTTTTATGACATTTGCACGTATTTCTAACATCTCAATTGCATATAAAAAG---------------------------TTTTT---------AA------------------------------------------------------------------T-------------------------------A--T-CTTTACC-CTAA-----CAAACATTTAACAT----ATAT-----------CTGTTGCAGAAAACTGAGCTCCTTGGGCAACGGCGA---ACCTTTCGATGCTTTAGACCCTG |
| droSec2 | scaffold_22:177884-178241 + | dse_1081 | CTGGGCGGCAAGCGGGCGCTCACCACATCCTGGAGCGTAGAGAAAAAGGGGTGGGTTTGTATACTAAACTATAGTTATTGTCTATATGCACATTAT-AA--------------------------GATGAAATTCGCTTGATTGTA----AATACTGCCACTG-GCCA-CAAGCATAAACATAAACATAAACATAATTTCTCCTTATGACTTCACATTTCTTCCTG----GAAGTCA------------------TG-AATGTTAACTTTTATGACATTTGCACGTATTTCTAACATCTCAATTGCATATAAAAAG---------------------------TTATT---------AA------------------------------------------------------------------T-------------------------------A--T-CTTTACC-CTAA-----CAAATATTTAACAT----ATAT-----------CTGTTGCAGAAAACTGAGCTCCTTGGGCAACGGCGA---ACCTTTCGATGCTTTAGACCCTG |
| dm3 | chr3R:23535040-23535397 + | CTGGGCGGCAAGCGGGCGCTCACCACATCCTGGAGCGTAGAGAAAAAGGGGTGGGTTTGTATACTAAACTATAGTTATTCTCTATGTGCACATTAT-AA--------------------------GATGAAATTCGCTTAATTGTA----AATACTGCCACTG-GCCA-CAAGCATAAACGTAAACATAAGCATAATTTCTCTTTATGACTTCACATTTCTTCCTG----GAAGTCA------------------TG-AATGTTAACTTTTATGACATTTGTACGTATTTCTAACATTTCAATTGCATATAAAAAG---------------------------TTTTT---------GA------------------------------------------------------------------T-------------------------------A--T-CTTTACC-CTAA-----CAAACATTAAATAT----ATAT-----------CTATTGCAGAAAACTGAGCTCCTTGGGCAATGGCGA---ACCTTTCGATGCTTTAGACGCCG | |
| droEre2 | scaffold_4820:4500249-4500600 - | CTGGGCGGCAAGCGGGCGCTCACCACATCCTGGAGCGTAGAGAAGAAGGGGTGGGTTTGTACTTTAAACTATAGTTATTGTCTGTATGCACATTAT-AA--------------------------GATGGAATTTGCTTTATTGTG----AATACTACCTCCG-GGCA-AAAGCAAAAGCATAAAC------ATAAGTTCTGTTTATGACATAACCTTTGAATGCG----GCAGTCA------------------GG-AAAGTTAACGTTTTTGGTATTTGCAACTATTCCTAACATCTCAATTGCATATAAACAG---------------------------TTTTT---------AA------------------------------------------------------------------C-------------------------------G--T-ATTTAGC-TAAA-----CAAACATTAAATAT----ATAT-----------ATCTTGCAGAAAACTCGGCTCCTTGGGCAATGGCGA---GCCATTCGATGCTTTAGACGCTG | |
| droYak3 | 3R:22951488-22951833 - | CTGGGCGGCAAGCGGGCGCTTACTACATCCTGGAGCGTAGAGAAAAAGGGGTGGGTTTGTATTTTAAACTGCATTGGTTATCTATATGCACATTAT-AA--------------------------GATGACTCTTATTTAATTGTAAATTAATTGTGCCCCTG-GGCA-CAAGCATAAAC------------ATAAATTCTGTTTATGACATCACATTTGAACCGG----GAAGTCA------------------TA-AAAGTTTACTTTTATGATATTTGCCAGTATTCCTAACATCTCAATTGCATATAAATAG---------------------------TCTTT---------AA------------------------------------------------------------------T--------------------------------------TTAGC-TAAG-----AAAACTTTAAATAA----ATAT-----------CTTTTGCAGAAAACTTGGCTCCTTGGGCAATGGCGA---GCCTTTCGATGCTTTAGACGCTG | |
| droEug1 | scf7180000409770:1020489-1020825 + | TTGGGTGGCAAGCGGGCGCTCACCACATCCTGGAGTGTAGAGAAAAAGGAGTGGGTGTACATTTTAAAAACTTGATATTGTTTATATGCA-ATTAA-AAGATTTTCCATTAGCG----------------------------------------ATGCCGTTG-GGAATAGAGCATAG---TAAACTGAATC---AA--TTCTTTATGATTTTGCATTTGCTTGTGTATAGAAGATA--------------GTAATAAAATGTTGGCAT----------------------------TTATTTTGCATATAAGCAA---------------------------TATAT---------AA------------------------------------------------------------------A-------------------------------T--G-TATTACA-CTAATAAGAAAAACATTAAATAA----TAATCTATAATCTATTTTTTGCAGAAAACTTGGCTCCATGGGCAATGGTGA---ACCTTTCGATGCTTTAAACGCTG | |
| droBia1 | scf7180000302075:1382174-1382475 - | CTGGGCGGCAAGAGGGCTCTGACCACCTCATGGAGCGTAGAAAAAAAGGAGTGAGTGGCTTATTTAGATACTGAGTACTGTTTATACGCAA-TCTG-TAAATCTCCCTTTACCAATTACTGTTTAGTTG------------GTGCT----AATATTGCCTATG-GGTA---------------AACTAAATC---GA--ATCCTTATGATTG-----------------------------------------------------------------------------CCTAACATCTTTCTTGCATATAAACAG---------------------------TATTA---------AA------------------------------------------------------------------T-------------------------------T--G-ATTCGCA-TTAA-----CAAATATTAAATAA----AAATCT-----C--TGTTTTGCAGCAAACTGGCCTCCCTGGGCAATGGCGA---ACCTTTCGATGCTTTGGACGCTG | |
| droTak1 | scf7180000415672:106550-106889 - | CTGGGTGGGAAGCGGGCGCTTACAACCTCATGGAGTGTGGAGAAAAAGGAGTGGGTGGCTATTTTAAAAACTAATTATTGTTTATATGCGA-TCTGTTTGATTTTCTATTAGCAATTATTGTTTAGATGGTAATTTATTAGGCGCA----AATATTGCTCCTGTGCCA-AAAACATAA---TGAACTCAATC---AA--TTCTTTATGATTTTAGATTTGTTGGCGCTTGAAAGA----------------------------TA------ATGATATT----------------------ATTGCATATAAACAG---------------------------TATTA---------GA------------------------------------------------------------------T-------------------------------A--T----------CAA-----CAAACATTAAATAAACCAATAT-----------TTATTACAGAAAACTTGGCTCCCTGGCCAATGGAGA---ACCATTCGATGCTTTAGACGCCG | |
| droEle1 | scf7180000491080:3433939-3434083 + | TTA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTAACAGCTCCGTTTCATAAATACAA---------------------------TATAAATTTTTATTTATTT---------------------------------------------------------------A-------------------------------T--T-TATTACA-TTAA-----CAAACATTAATTAG----GAATCC-----CTTCCTTTTCCAGCAAACTGGGCTCCTTGGGCAATGGCGA---ACCTTTCGACGCTTTGGACGCAG | |
| droRho1 | scf7180000779572:189349-189472 + | TATTCTTGAAAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTCCGTTGCGTAGAAAAAA---------------------------ATATC---------AA------------------------------------------------------------------C-------------------------------A--TTAATTACA-TTAA-----CTAATATTCC----------CT-----------TTCTTTCAGAAAACTGGGCTCCTTGGGAAATGGAGA---ACCTTTCGATGCTTTGGACGCAG | |
| droFic1 | scf7180000454055:570375-570477 - | CTAGGCGGCAAGCGAGCGCTAACCACCTCATGGAGCGTTGAAAAAAAAGAGTGGGTTCTTATTTTCAACCGGGAATATTGT-TGCATACAACTTAT-AG--------------------------TATCAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droKik1 | scf7180000302697:810839-810903 + | CTGGGCGGCAAGCGGGCCCTTACCACATCCTGGAGCGTCGAGAAGAAGGAGTAGGTTATTGCACT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13340:19154039-19154107 - | CTGGGCGGCAAGCGAGCACTCACCACCTCTTGGAGCGTGGAAAAAAAGGAGTAAGTTTTTATGTCAAAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBip1 | scf7180000395652:27867-27930 + | CTGGGCGGCAAGCGAGCCCTCACTACCTCCTGGAGCATGGAGAAAAAGGAGTAGGTTTTTAAAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| dp5 | 2:3894558-3894614 + | CTGGGTGGCAAGCGGGCCCTGACCACGTCCTGGAGTGTGGAAAAGAAGGAGTAGGTT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004902:3777322-3777411 + | TTTGCCATT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T-----TAATCATTTTGCTT----CATT-----------TTATTTTAGGAAATTAAGCTCCAAGGCGGGTGGCAAGCAACCTTTCGACAATCTAGAATCTG | |
| droVir3 | scaffold_12855:2070716-2070772 - | CTGGGCGGCAAGCGTGCGCTCAATACATCCTGGAGTGTGGAAAAGAAGGAGTAGGTT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droMoj3 | scaffold_6540:32367441-32367510 - | CTGGGCGGCAAGCGGGCGCTCACCACATCTTGGAGCGTGGAAAAGAAGGAGTGCGTATGCAGGGAAAACT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droGri2 | scaffold_14830:440973-441405 - | CTCGGCGGCAAGCGGGCCCTTACCACATCTTGGAGCGTGGAGAAGAAGGAGTATGTTCCACACACACAGACAAATTGTCTTT-------------TTCTGATACTCTATTCTCAA-----------ATGGAAA---ACAAGGCGT------GTATTGCACGTCTGTCA---------------------------------------GTTTTTCCCTCTGTCCGCT----TAAATTCGGAAATATTATATTGAACTTAAACGTTTTCAT----------------------------TTAATT------CAGGCGATGATGTAAAATAATTCAAATATTCTCAAATAATTTTTTATCAATCCATTTAGTTTTTGGAATTCTTTCCAATATCTTTCTAAAACTTTTCAAATATTGTCAAATGTCAGCTAACATTCACTGAGAACATAGAATCAGTAGACTCTAAACTACATCTGG-----CTAATATTTCCC------TTAT-----------TTTTTCTAGCAAATTGAGCTCCACGGC------AGA---GCCCTTCGATTCGTTGTGCGCGG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 09:50 PM