ID:dsi_10568 |
Coordinate:3l:7693327-7693476 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
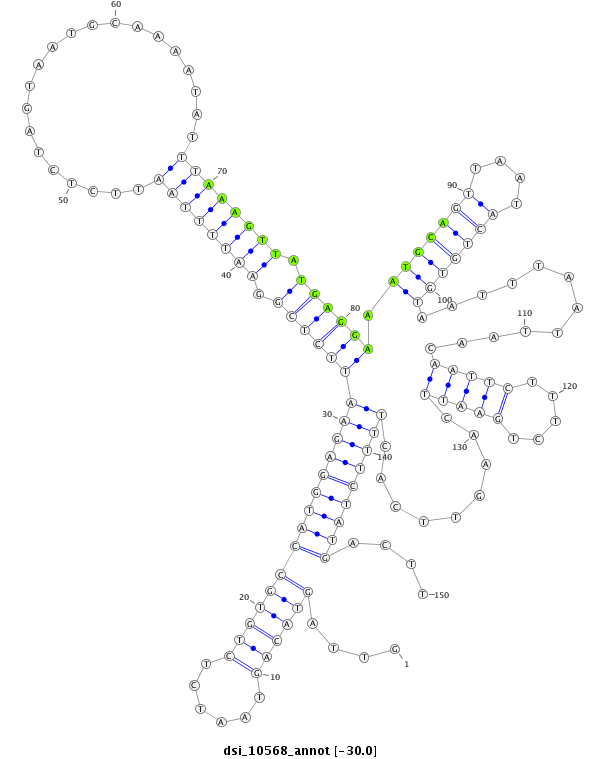
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -29.7 | -29.6 | -29.5 |
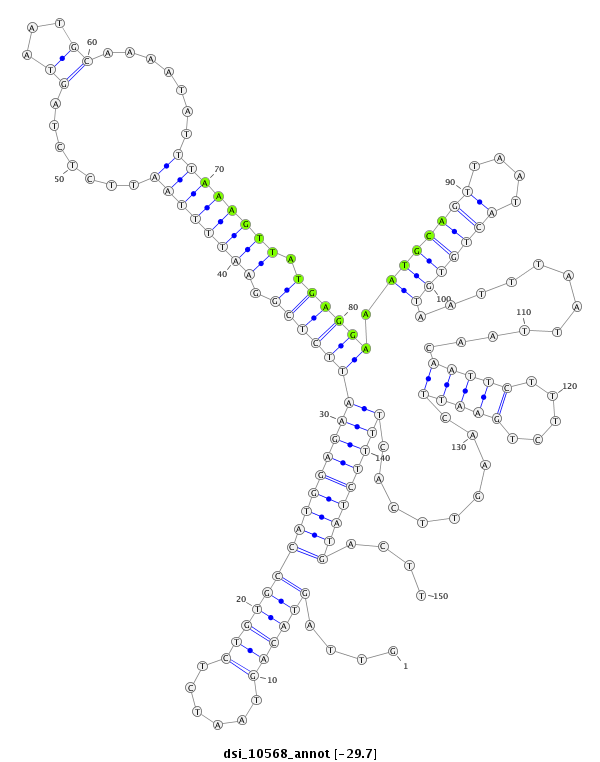 |
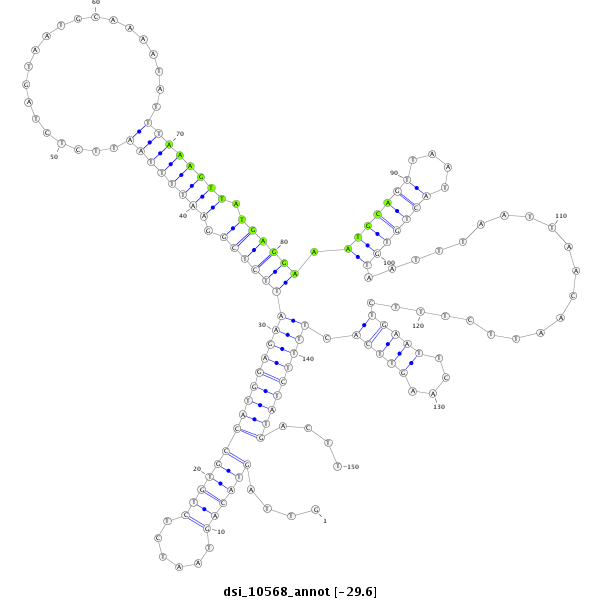 |
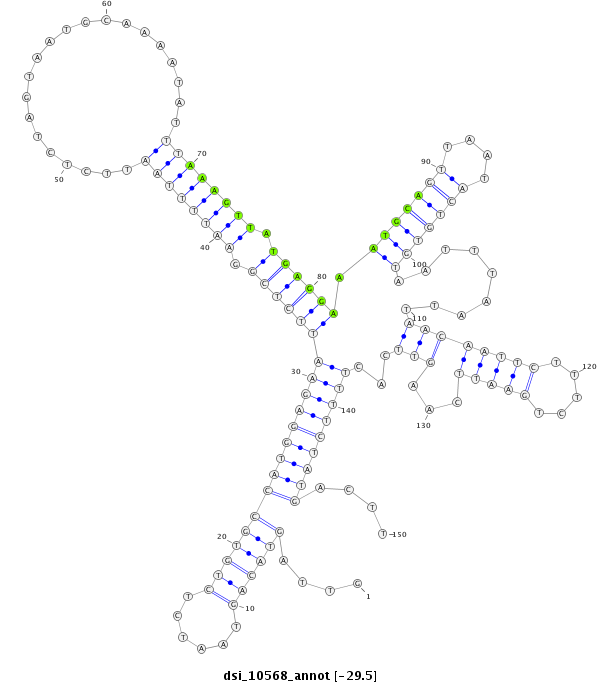 |
CDS [3l_7692874_7693326_+]; exon [3l_7692865_7693326_+]; intron [3l_7693327_7710111_+]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TCAATCTCGTCTGGCCAATGCAACCGACACATCAGCCAGCACAGCAGCAGGTTAGTACAGTAATCTCTGTGCCATGGAGAATTCTCGGAATTTTAATTCTCTAGTAATGCAAAATATTTAAAGTTATGAGGAAATGCAGTTAATACTGTGTAATTTAATTAACAATTCTTTCTGAATTCAAGTTCACTTTTCTATGACTTGGAATTCCCTGAAGTTCTGTTACGCGAGTGCCCACTGTATCACTTTATGC **************************************************....((((((......))))))(((((((((((((((.((((((((.....................)))))))).)))))).(((((((....)))))))............(((((.....))))).........)))))))))....************************************************** |
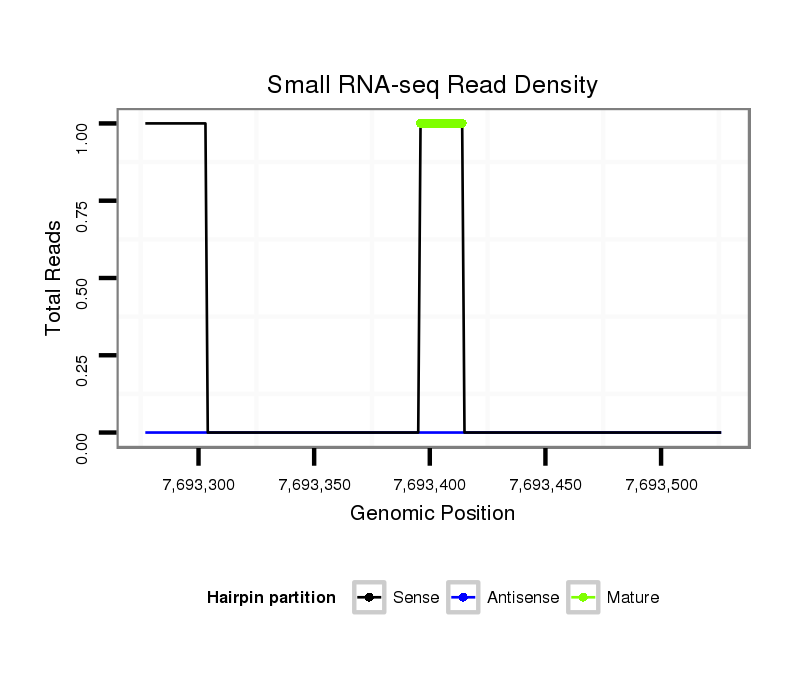
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
M024 male body |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TCAATCTCGTCTGGCCAATGCAACCGA............................................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................AAAGTTATGAGGAAATGCA................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................ACTTGGAATTCCCAGTA..................................... | 17 | 2 | 20 | 0.55 | 11 | 9 | 0 | 1 | 0 | 0 | 1 |
| ....................................................................................................................................................................................................ACTTGGAATTCCCTG....................................... | 15 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................ACTTGGAATTCCCGGAAC.................................... | 18 | 2 | 6 | 0.33 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................ACTTGGAATTCCCAGTAG.................................... | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................AACTTGGAATTCCCAGTAGT................................... | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................TACGACTTGGAATTCCCA........................................ | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................GACTTGGAATTCCCA........................................ | 15 | 1 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................AAGACTTGGAATTCCCA........................................ | 17 | 2 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................CAACTCTGTGCAATGGAGA.......................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........TCTGGCCAATGAAAC.................................................................................................................................................................................................................................. | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
|
AGTTAGAGCAGACCGGTTACGTTGGCTGTGTAGTCGGTCGTGTCGTCGTCCAATCATGTCATTAGAGACACGGTACCTCTTAAGAGCCTTAAAATTAAGAGATCATTACGTTTTATAAATTTCAATACTCCTTTACGTCAATTATGACACATTAAATTAATTGTTAAGAAAGACTTAAGTTCAAGTGAAAAGATACTGAACCTTAAGGGACTTCAAGACAATGCGCTCACGGGTGACATAGTGAAATACG
**************************************************....((((((......))))))(((((((((((((((.((((((((.....................)))))))).)))))).(((((((....)))))))............(((((.....))))).........)))))))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................AAGTTCAAGTGAAAATATA....................................................... | 19 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| AGTTAGAGCACACCGAT......................................................................................................................................................................................................................................... | 17 | 2 | 6 | 0.33 | 2 | 0 | 1 | 1 |
| AGTTAGAGCACACCGATT........................................................................................................................................................................................................................................ | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 |
| AGTTAGAGCACACCG........................................................................................................................................................................................................................................... | 15 | 1 | 10 | 0.20 | 2 | 2 | 0 | 0 |
| .....................................................................................................................................................................................CAAGTGACAAGATTCTG.................................................... | 17 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 |
| ......................................................................TGGTACCTCTTAAGATCGT................................................................................................................................................................. | 19 | 3 | 14 | 0.14 | 2 | 0 | 0 | 2 |
| ....................................................................................................................................................................................................................................TCAGGTGACATAGTG....... | 15 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 |
Generated: 04/24/2015 at 10:27 AM