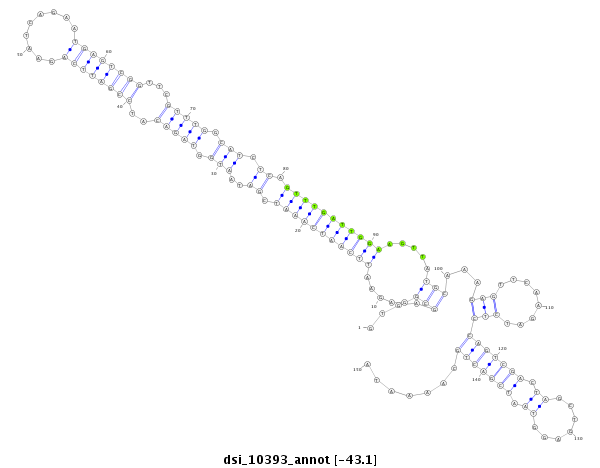
ID:dsi_10393 |
Coordinate:3l:16317871-16318020 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -42.5 | -42.1 |
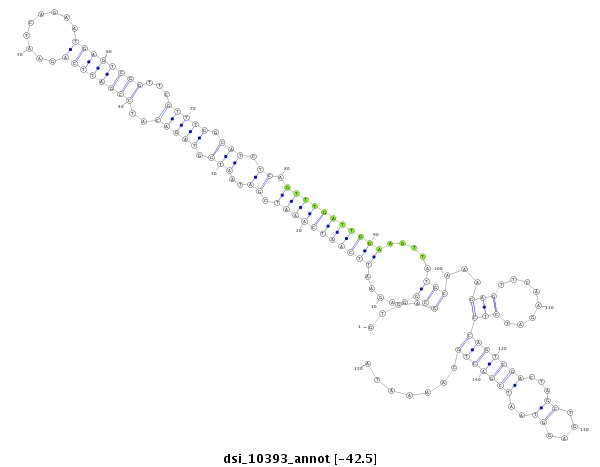 |
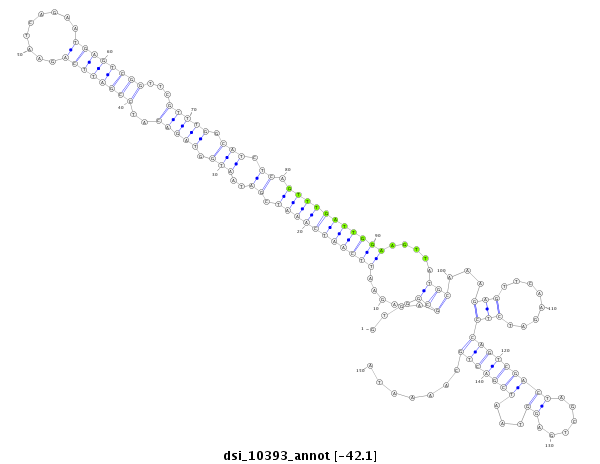 |
exon [3l_16317669_16317870_+]; CDS [3l_16317669_16317870_+]; intron [3l_16317871_16320455_+]
No Repeatable elements found
| mature | star |
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AAATAGTCGATCAATCCAATGGCGATGTCACGGCGGACAGCTATCATCAGGTGAGCGGAGAATTCAATCAAATCGATAATGGTAGACATCCGATTCAGAATCAGAATGAGTCGGTTCGTTTGGCATCTCAGTTTGATTGGAAGTTATGCAAAGAGTTCAAGATCTCCAGTCGACTAGCTGAGGTAATCGACTGCAAAATATGTAGGCAACTGGCAATTGATGCACTCCGATGGCTATTAGTTACACAGCT **************************************************....(((.....(((((((((((.((..(((.(((((..((((((((.........))))))))...))))).))).)).))))))))))).....)))...(((........)))(((((((.((.......)).))))))).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR1275483 Male prepupae |
SRR553486 Makindu_3 day-old ovaries |
SRR902009 testis |
SRR553485 Chicharo_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................GTTTGATTGGAAGTT......................................................................................................... | 15 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................TCTACAGTCGACGATCTGA..................................................................... | 19 | 3 | 16 | 0.31 | 5 | 0 | 5 | 0 | 0 | 0 | 0 |
| ...............CCAAACGCGATGTTACGGCG....................................................................................................................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................AGCGATGCACTCCGAAGGCT............... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................AGAGTTCTAGTTCTACAGTC............................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................TCAAGTTCTACAGTCGACG........................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................AAGCGTCGGTTCGCTTGGC.............................................................................................................................. | 19 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TGAGTCGGGTTGTTTGG............................................................................................................................... | 17 | 2 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................TGGTAGACATCGCATT........................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................................................................................TCGACGAGCTGAGGT.................................................................. | 15 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
|
TTTATCAGCTAGTTAGGTTACCGCTACAGTGCCGCCTGTCGATAGTAGTCCACTCGCCTCTTAAGTTAGTTTAGCTATTACCATCTGTAGGCTAAGTCTTAGTCTTACTCAGCCAAGCAAACCGTAGAGTCAAACTAACCTTCAATACGTTTCTCAAGTTCTAGAGGTCAGCTGATCGACTCCATTAGCTGACGTTTTATACATCCGTTGACCGTTAACTACGTGAGGCTACCGATAATCAATGTGTCGA
**************************************************....(((.....(((((((((((.((..(((.(((((..((((((((.........))))))))...))))).))).)).))))))))))).....)))...(((........)))(((((((.((.......)).))))))).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|
| ...................................................................................................TAGTCTTACTCACCC........................................................................................................................................ | 15 | 1 | 7 | 0.14 | 1 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................................CCCCCGATAATGAATGTGT... | 19 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 |
| .........................................................................................................................................................................AGATGGTTGACTCCATTAG.............................................................. | 19 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 |
| .............................................................................................................................AGGGTCAAACTAACC.............................................................................................................. | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 |
Generated: 04/24/2015 at 04:11 AM