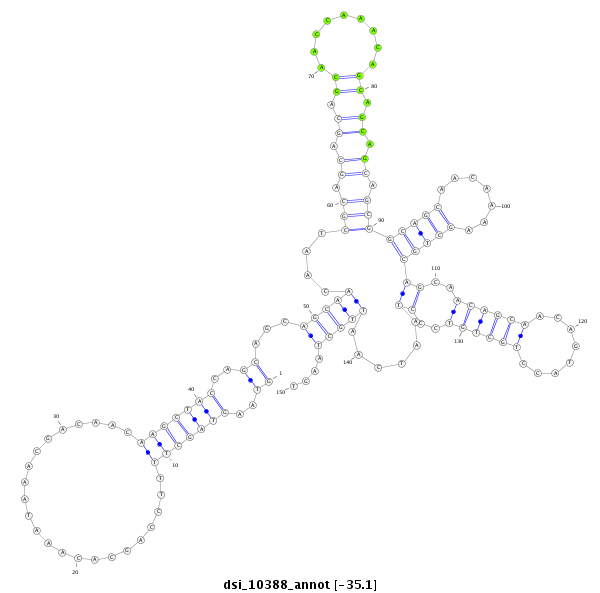
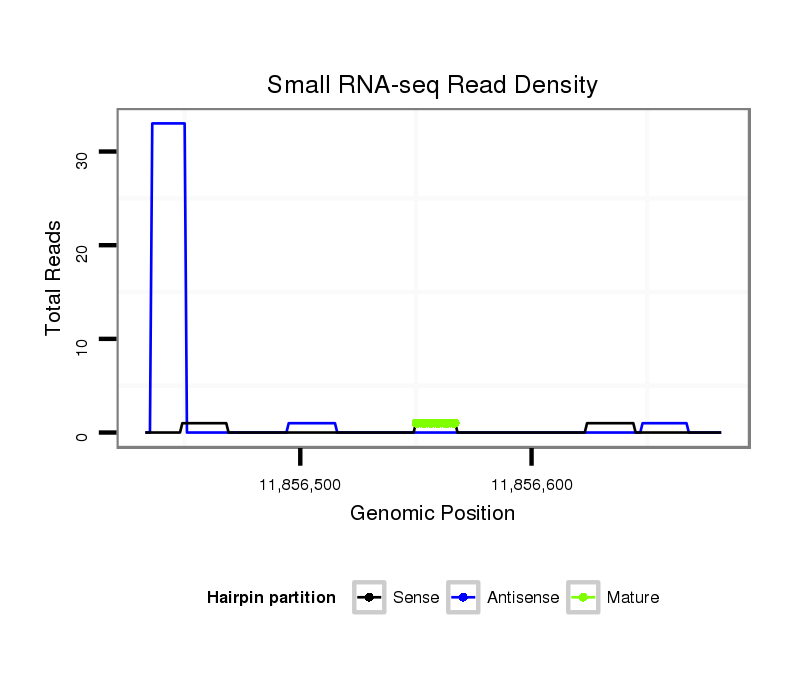
| TTGCAGCA---------------------------ACAA--CAGCAACAGCAGCAACAATGGCAGCAACAACA---ACAGCAA-------------------------CAACAACTGCAGCAGCAAC----AACAACA-----GCAGCAACAACAACTGCAGCAGCAACAATTGCAGCAACAACAACAACAGCAGCAACA------ACAACAGC-AGCAGCAACAACAGCA--------------------TCAGCAACAGCAACA---G--------------------------------------------------CA---GCA---------ACGAATGCAAC-------AGCAACAGCAAC----------------------------------------------------AGCAGCAACAATTGCAGC---AACAACAACA-----------ACAGCAACAACAACAAC | Size | Hit Count | Total Norm | Total | M020
Head | M045
Female-body | V117
Male-body | V118
Embryo | V119
Head |
| ......................................................AACAATGGCAGCAACAACA---ACAG.............................................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 20 | 0.70 | 14 | 0 | 14 | 0 | 0 | 0 |
| .......A---------------------------ACAA--CAGCAACAGCAGCAACAATGG................................................................................................................................................................................................................................................................................................................................................................................................................................ | 26 | 20 | 0.35 | 7 | 0 | 7 | 0 | 0 | 0 |
| .........................................................................................................................................................................AATTGCAGCAACAACAACAACAGC............................................................................................................................................................................................................................................................................................. | 24 | 10 | 0.20 | 2 | 0 | 2 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................AATTGCAGC---AACAACAACA-----------ACAGC............ | 24 | 10 | 0.20 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................TGCAGC---AACAACAACA-----------ACAGCAACAACA..... | 28 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................GCAGCAACA------ACAACAGC-A...................................................................................................................................................................................................................................................................... | 18 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 |
| ....AGCA---------------------------ACAA--CAGCAACAGCAGCAA...................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................AACAGCAAC----------------------------------------------------AGCAGCAACA................................................. | 19 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................CA---------ACGAATGCAAC-------AGCAACAGCAAC----------------------------------------------------........................................................... | 25 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................TGCAGC---AACAACAACA-----------ACAGCAACA........ | 25 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................TCAGCAACAGCAACA---G--------------------------------------------------CA---GCA---------ACGAA........................................................................................................................................ | 26 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................AATTGCAGC---AACAACAACA-----------ACAG............. | 23 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......A---------------------------ACAA--CAGCAACAGCAGCAACAA................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................ACAACAGC-AGCAGCAACAAC........................................................................................................................................................................................................................................................... | 20 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 |
| ......................................................................................................................................................CAACAACTGCAGCAGCAACAATTGC............................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ......................................................AACAATGGCAGCAACAACA---ACAGC............................................................................................................................................................................................................................................................................................................................................................................................................. | 24 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ......................................................................................................................................................................AACAATTGCAGCAACAACAACAACAGC............................................................................................................................................................................................................................................................................................. | 27 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| .TGCAGCA---------------------------ACAA--CAGCAACAGCAG......................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................AACAATTGCAGC---AACAACAACA-----------ACAGC............ | 27 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....AGCA---------------------------ACAA--CAGCAACAGCAG......................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ..........................................................................................................................GCAAC----AACAACA-----GCAGCAACA...................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| .........................................................................................................................................................................AATTGCAGCAACAACAACAACAG.............................................................................................................................................................................................................................................................................................. | 23 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................AGCAACAGCAACA---G--------------------------------------------------CA---GCA---------ACG.......................................................................................................................................... | 22 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................CAACAACAGCA--------------------TCAGCAACAGCAACA---G--------------------------------------------------.............................................................................................................................................................. | 27 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................CAACAGCA--------------------TCAGCAACAGCAACA---G--------------------------------------------------CA---G........................................................................................................................................................ | 27 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................CAACAGC-AGCAGCAACAACAGCA--------------------TCAG............................................................................................................................................................................................................................... | 27 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................AATTGCAGCAACAACAACA.................................................................................................................................................................................................................................................................................................. | 19 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................AATTGCAGC---AACAACAACA-----------................. | 19 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................CAACAACAGCA--------------------TCAGCAACAGC........................................................................................................................................................................................................................ | 22 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................TGCAGCAGCAAC----AACAACA-----GCAGCAACA...................................................................................................................................................................................................................................................................................................................................... | 28 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................AACAACTGCAGCAGCAAC----AACAACA-----............................................................................................................................................................................................................................................................................................................................................... | 25 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................AGCAACAGCAAC----------------------------------------------------AGCAGCAACAATT.............................................. | 25 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................TGCAAC-------AGCAACAGCAAC----------------------------------------------------AGCAGC..................................................... | 24 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................AGCA--------------------TCAGCAACAGCAACA---G--------------------------------------------------CA---G........................................................................................................................................................ | 23 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................CAACAGCA--------------------TCAGCAACAGC........................................................................................................................................................................................................................ | 19 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................A--------------------TCAGCAACAGCAACA---G--------------------------------------------------CA---GCA---------AC........................................................................................................................................... | 24 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................CAGCAGCAACAATGGCAGCAA.......................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................AACAACAACTGCAGCAGCAAC..................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................AATGGCAGCAACAACA---ACAGCAA-------------------------CAAC.............................................................................................................................................................................................................................................................................................................................................................................. | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................AACAACTGCAGCAGCAACAATTG................................................................................................................................................................................................................................................................................................................ | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................GGCAGCAACAACA---ACAGCAA-------------------------CAA............................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................AATGGCAGCAACAACA---ACAGC............................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................CA---ACAGCAA-------------------------CAACAACTGCAG...................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................ACAATGGCAGCAACAACA---ACAG.............................................................................................................................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................AACAACA---ACAGCAA-------------------------CAACAACTGCAGC..................................................................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................AA-------------------------CAACAACTGCAGCAGCAAC----........................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................CAAC-------AGCAACAGCAAC----------------------------------------------------AGCAG...................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .TGCAGCA---------------------------ACAA--CAGCAACAGCA.......................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................CAACA---ACAGCAA-------------------------CAACAACTGCAGC..................................................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................AGCAACAACA---ACAGCAA-------------------------CAACAACTGCAG...................................................................................................................................................................................................................................................................................................................................................................... | 29 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................GGCAGCAACAACA---ACAGCAA-------------------------C................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................TGGCAGCAACAACA---ACAGCAA-------------------------CAACAA............................................................................................................................................................................................................................................................................................................................................................................ | 27 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................AGCAACAATGGCAGCAACAACA---ACA............................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................A---ACAGCAA-------------------------CAACAACTGCAGCAGCAAC----........................................................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................CAACA------ACAACAGC-AGCAGC................................................................................................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................AACAATTGCAGC---AACAACAACA-----------G................ | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................ACAGCAA-------------------------CAACAACTGCAGCA.................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................A------ACAACAGC-AGCAGCAACAA............................................................................................................................................................................................................................................................ | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................AGCAA-------------------------CAACAACTGCAGCAGCAAC----A.......................................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .TGCAGCA---------------------------ACAA--CAGCAACAGCAGC........................................................................................................................................................................................................................................................................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................AATGGCAGCAACAACA---ACAGCAA-------------------------CA................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................CAA--CAGCAACAGCAGCAACAATGG................................................................................................................................................................................................................................................................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................CAACAACTGCAGCAGCAACAATTGCAGC............................................................................................................................................................................................................................................................................................................ | 28 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................ACAGCAGCAACA------ACAACAGC-A...................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ......CA---------------------------ACAA--CAGCAACAGCAGCAAC..................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................TGGCAGCAACAACA---ACAGCAA-------------------------CAACAAC........................................................................................................................................................................................................................................................................................................................................................................... | 28 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................AGCAGCAACAATGGCAGCAACAACA---.................................................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| TTGCAGCA---------------------------ACAA--CAGCAACAGCAG......................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................................................AGCAACAGCAACA---G--------------------------------------------------CA---GCA---------............................................................................................................................................. | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................A---ACAGCAA-------------------------CAACAACTGCAGCAGC.................................................................................................................................................................................................................................................................................................................................................................. | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................AACAATTGCAGCAACAACAACAG................................................................................................................................................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................AGCAAC----------------------------------------------------AGCAGCAACAATTG............................................. | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................AACAACA-----------ACAGCAACAACAACA.. | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................CAACAATGGCAGCAACAACA---ACAG.............................................................................................................................................................................................................................................................................................................................................................................................................. | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| TTGCAGCA---------------------------ACAA--CAGCAACAGCAGC........................................................................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................AACAACA---ACAGCAA-------------------------CAACAACT.......................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................AGCAA-------------------------CAACAACTGCAGCAGCAA................................................................................................................................................................................................................................................................................................................................................................ | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................AGCAACAGCAGCAACAATGGCAG............................................................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................AACAACA---ACAGCAA-------------------------CAACAACTGC........................................................................................................................................................................................................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................TGGCAGCAACAACA---ACAGCAA-------------------------CAA............................................................................................................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................AAC----AACAACA-----GCAGCAACAAC.................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................ACAATGGCAGCAACAACA---ACAGCA............................................................................................................................................................................................................................................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................AACAACTGCAGCAGCAACAATT................................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................AAC-------AGCAACAGCAAC----------------------------------------------------AGCAGC..................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |