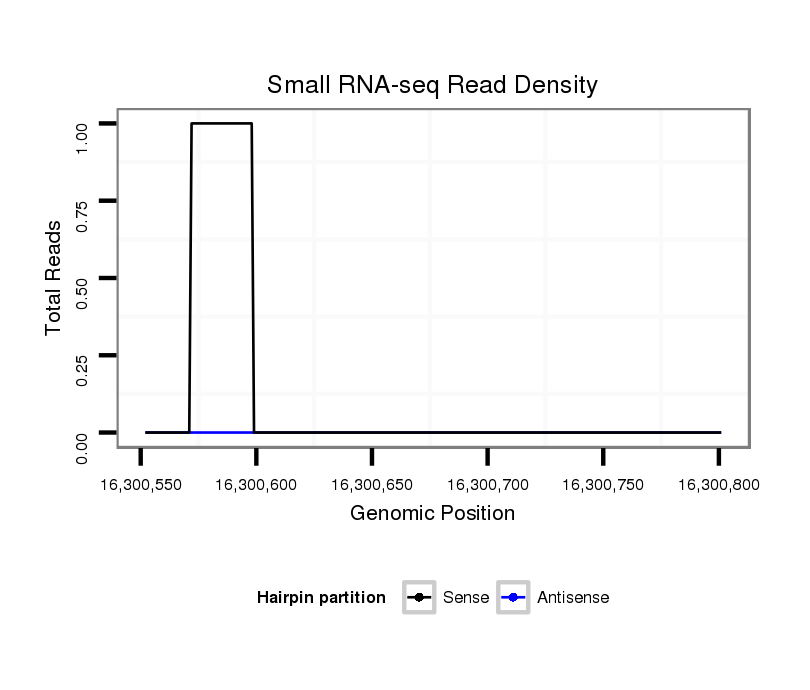
ID:dsi_10014 |
Coordinate:3l:16300602-16300751 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
CDS [3l_16300752_16300985_+]; exon [3l_16300752_16300985_+]; intron [3l_16300015_16300751_+]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TAGGGATCTTCGTTTGTGGATTTGTTGTCTTCGGAACTTATCGAAGATATAAGCTTTCTCTTCCATTTCTTCTGAGCTCTTTGAGTTTTTCTCCTGTTTTTGATTTTGTGCCAAATGCGAATTCGGAGTCTCCGCTTGGAGCTGCTCTCGTTGCTTGTTATCCCTGTCGTTTTCCTGATCCCTTCTGTTCCATTGTTCAGATGCGTTCACGGATTTGGATCCGCTAGGCACAGGACGCACCAGGCCGTAT **************************************************((((...................((((.((((((((....((((....((((..((((...))))..))))...)))).....))))))))..)))).....))))..................((((..................))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553485 Chicharo_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| ....................TTTGTTGTCTTCGGAACTTATCGAAGA........................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......CTTCGTTTGTGTATTTGGTAT.............................................................................................................................................................................................................................. | 21 | 3 | 6 | 0.67 | 4 | 0 | 4 | 0 | 0 | 0 |
| .....................TCGTTGTCTTCGGAAC..................................................................................................................................................................................................................... | 16 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................CTGTCGTATTCCGGATCGC.................................................................... | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........GTTTTTGGAGTTGTTGTCATC.......................................................................................................................................................................................................................... | 21 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................GGCTCAGGACGAACCTGGC..... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..............TGTGGATCTGTTGTCCT........................................................................................................................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
|
ATCCCTAGAAGCAAACACCTAAACAACAGAAGCCTTGAATAGCTTCTATATTCGAAAGAGAAGGTAAAGAAGACTCGAGAAACTCAAAAAGAGGACAAAAACTAAAACACGGTTTACGCTTAAGCCTCAGAGGCGAACCTCGACGAGAGCAACGAACAATAGGGACAGCAAAAGGACTAGGGAAGACAAGGTAACAAGTCTACGCAAGTGCCTAAACCTAGGCGATCCGTGTCCTGCGTGGTCCGGCATA
**************************************************((((...................((((.((((((((....((((....((((..((((...))))..))))...)))).....))))))))..)))).....))))..................((((..................))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
O001 Testis |
SRR618934 dsim w501 ovaries |
M025 embryo |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................AGCACCTCAAAAAGAAGACAAA....................................................................................................................................................... | 22 | 3 | 8 | 0.25 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................AGCAATAGCACTAGGGCAGAC............................................................... | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................CAAGTGCCTAAACGTA.............................. | 16 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................AGAAAATCAAAAAGAGGTC.......................................................................................................................................................... | 19 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........CAAACACAGAAACAACAGA............................................................................................................................................................................................................................ | 19 | 2 | 16 | 0.13 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................GGCGAATCTCAACCAGAGC.................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................TAGGACTAGGGAAGAGTAG............................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................AAAGGACGGGGGATGACAA............................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................................................................................................................ATGCACGTGCCTAAAC................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
Generated: 04/24/2015 at 04:07 AM